Plot Modifications: Overplotting Contours, Velocities, Particles, and More¶
Adding callbacks to plots¶
After a plot is generated using the standard tools (e.g. SlicePlot,
ProjectionPlot, etc.), it can be annotated with any number of callbacks
before being saved to disk. These callbacks can modify the plots by adding
lines, text, markers, streamlines, velocity vectors, contours, and more.
Callbacks can be applied to plots created with
SlicePlot,
ProjectionPlot,
AxisAlignedSlicePlot,
AxisAlignedProjectionPlot,
OffAxisSlicePlot, or
OffAxisProjectionPlot, by calling
one of the annotate_ methods that hang off of the plot object.
The annotate_ methods are dynamically generated based on the list
of available callbacks. For example:
slc = SlicePlot(ds, "x", ("gas", "density"))
slc.annotate_title("This is a Density plot")
would add the TitleCallback() to
the plot object. All of the callbacks listed below are available via
similar annotate_ functions.
To clear one or more annotations from an existing plot, see the clear_annotations function.
For a brief demonstration of a few of these callbacks in action together, see the cookbook recipe: Annotating Plots to Include Lines, Text, Shapes, etc..
Also note that new annotate_ methods can be defined without modifying yt’s
source code, see Extending annotations methods.
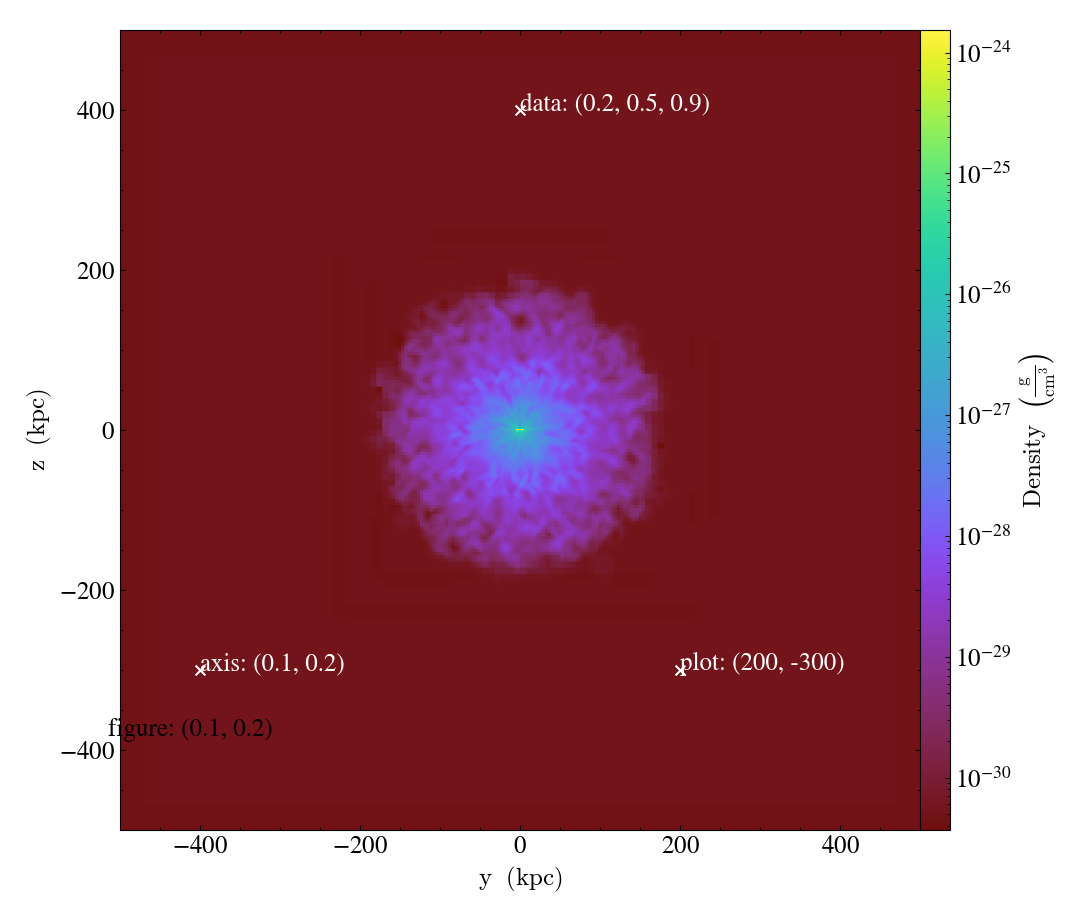
Coordinate Systems in Callbacks¶
Many of the callbacks (e.g.
TextLabelCallback) are specified
to occur at user-defined coordinate locations (like where to place a marker
or text on the plot). There are several different coordinate systems used
to identify these locations. These coordinate systems can be specified with
the coord_system keyword in the relevant callback, which is by default
set to data. The valid coordinate systems are:
data– the 3D dataset coordinates
plot– the 2D coordinates defined by the actual plot limits
axis– the MPL axis coordinates: (0,0) is lower left; (1,1) is upper right
figure– the MPL figure coordinates: (0,0) is lower left, (1,1) is upper right
Here we will demonstrate these different coordinate systems for an projection of the x-plane (i.e. with axes in the y and z directions):
import yt
ds = yt.load("IsolatedGalaxy/galaxy0030/galaxy0030")
s = yt.SlicePlot(ds, "x", ("gas", "density"))
s.set_axes_unit("kpc")
# Plot marker and text in data coords
s.annotate_marker((0.2, 0.5, 0.9), coord_system="data")
s.annotate_text((0.2, 0.5, 0.9), "data: (0.2, 0.5, 0.9)", coord_system="data")
# Plot marker and text in plot coords
s.annotate_marker((200, -300), coord_system="plot")
s.annotate_text((200, -300), "plot: (200, -300)", coord_system="plot")
# Plot marker and text in axis coords
s.annotate_marker((0.1, 0.2), coord_system="axis")
s.annotate_text((0.1, 0.2), "axis: (0.1, 0.2)", coord_system="axis")
# Plot marker and text in figure coords
# N.B. marker will not render outside of axis bounds
s.annotate_marker((0.1, 0.2), coord_system="figure", color="black")
s.annotate_text(
(0.1, 0.2),
"figure: (0.1, 0.2)",
coord_system="figure",
text_args={"color": "black"},
)
s.save()
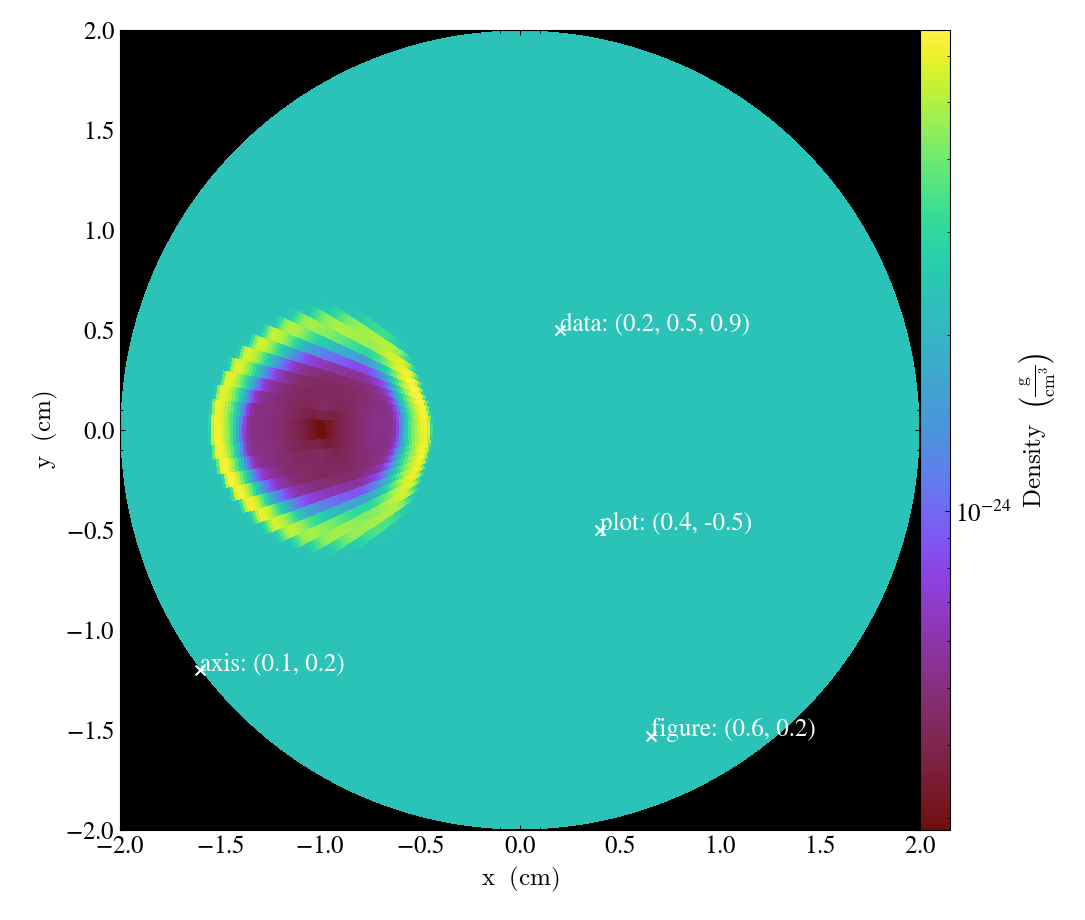
Note that for non-cartesian geometries and coord_system="data", the coordinates
are still interpreted in the corresponding cartesian system. For instance using a polar
dataset from AMRVAC :
import yt
ds = yt.load("amrvac/bw_polar_2D0000.dat")
s = yt.plot_2d(ds, ("gas", "density"))
s.set_background_color("density", "black")
# Plot marker and text in data coords
s.annotate_marker((0.2, 0.5, 0.9), coord_system="data")
s.annotate_text((0.2, 0.5, 0.9), "data: (0.2, 0.5, 0.9)", coord_system="data")
# Plot marker and text in plot coords
s.annotate_marker((0.4, -0.5), coord_system="plot")
s.annotate_text((0.4, -0.5), "plot: (0.4, -0.5)", coord_system="plot")
# Plot marker and text in axis coords
s.annotate_marker((0.1, 0.2), coord_system="axis")
s.annotate_text((0.1, 0.2), "axis: (0.1, 0.2)", coord_system="axis")
# Plot marker and text in figure coords
# N.B. marker will not render outside of axis bounds
s.annotate_marker((0.6, 0.2), coord_system="figure")
s.annotate_text((0.6, 0.2), "figure: (0.6, 0.2)", coord_system="figure")
s.save()
Available Callbacks¶
The underlying functions are more thoroughly documented in Callback List.
Clear Callbacks (Some or All)¶
- clear_annotations(index=None)¶
This function will clear previous annotations (callbacks) in the plot. If no index is provided, it will clear all annotations to the plot. If an index is provided, it will clear only the Nth annotation to the plot. Note that the index goes from 0..N, and you can specify the index of the last added annotation as -1.
(This is a proxy for
clear_annotations().)
import yt
ds = yt.load("IsolatedGalaxy/galaxy0030/galaxy0030")
p = yt.SlicePlot(ds, "z", ("gas", "density"), center="c", width=(20, "kpc"))
p.annotate_scale()
p.annotate_timestamp()
# Oops, I didn't want any of that.
p.clear_annotations()
p.save()
List Currently Applied Callbacks¶
- list_annotations()¶
This function will print a list of each of the currently applied callbacks together with their index. The index can be used with clear_annotations() function to remove a specific callback.
(This is a proxy for
list_annotations().)
import yt
ds = yt.load("IsolatedGalaxy/galaxy0030/galaxy0030")
p = yt.SlicePlot(ds, "z", ("gas", "density"), center="c", width=(20, "kpc"))
p.annotate_scale()
p.annotate_timestamp()
p.list_annotations()
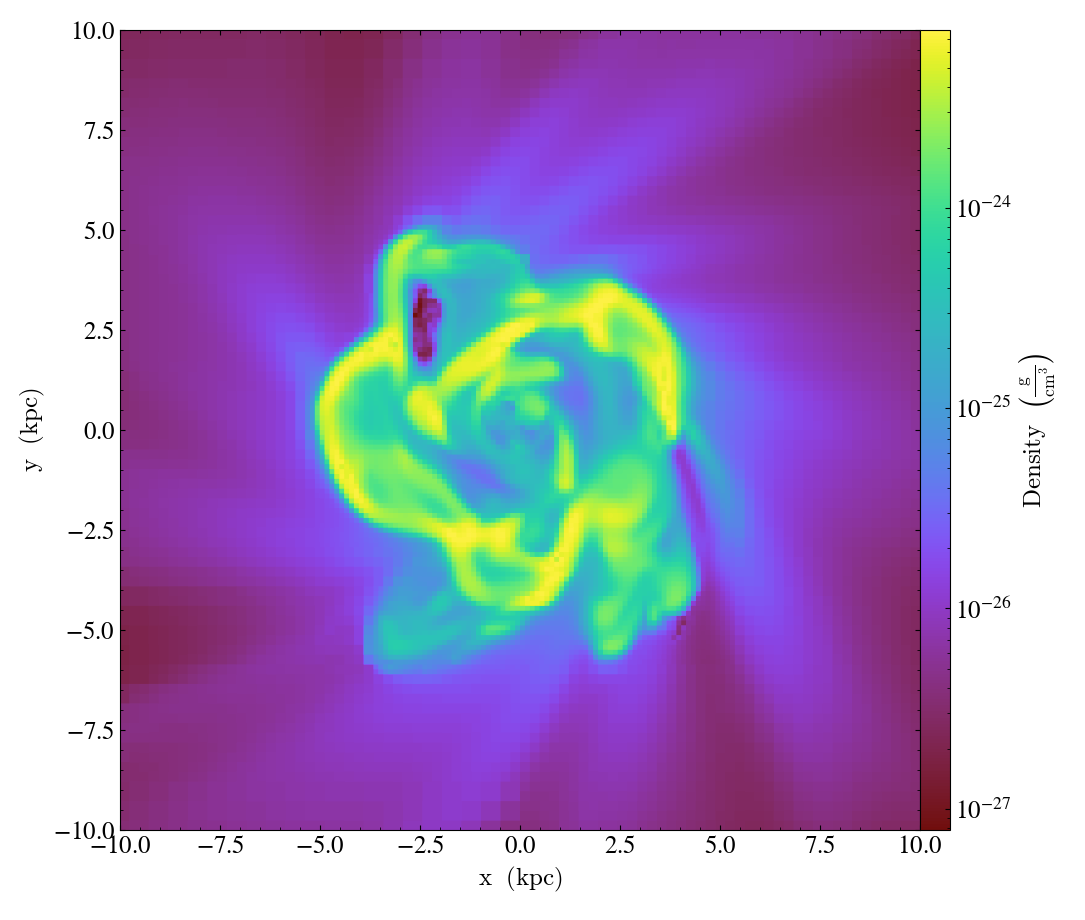
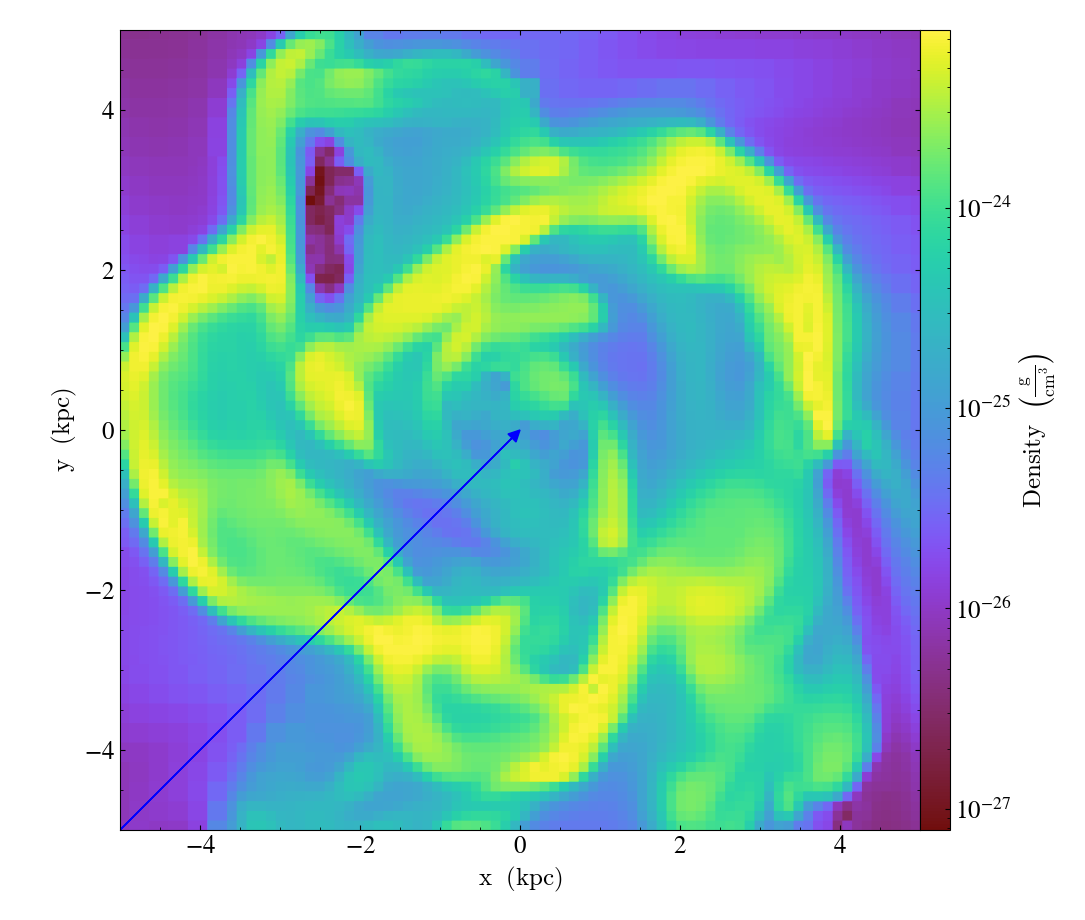
Overplot Arrow¶
- annotate_arrow(self, pos, length=0.03, coord_system='data', **kwargs)¶
(This is a proxy for
ArrowCallback.)Overplot an arrow pointing at a position for highlighting a specific feature. Arrow points from lower left to the designated position with arrow length “length”.
import yt
ds = yt.load("IsolatedGalaxy/galaxy0030/galaxy0030")
slc = yt.SlicePlot(ds, "z", ("gas", "density"), width=(10, "kpc"), center="c")
slc.annotate_arrow((0.5, 0.5, 0.5), length=0.06, color="blue")
slc.save()
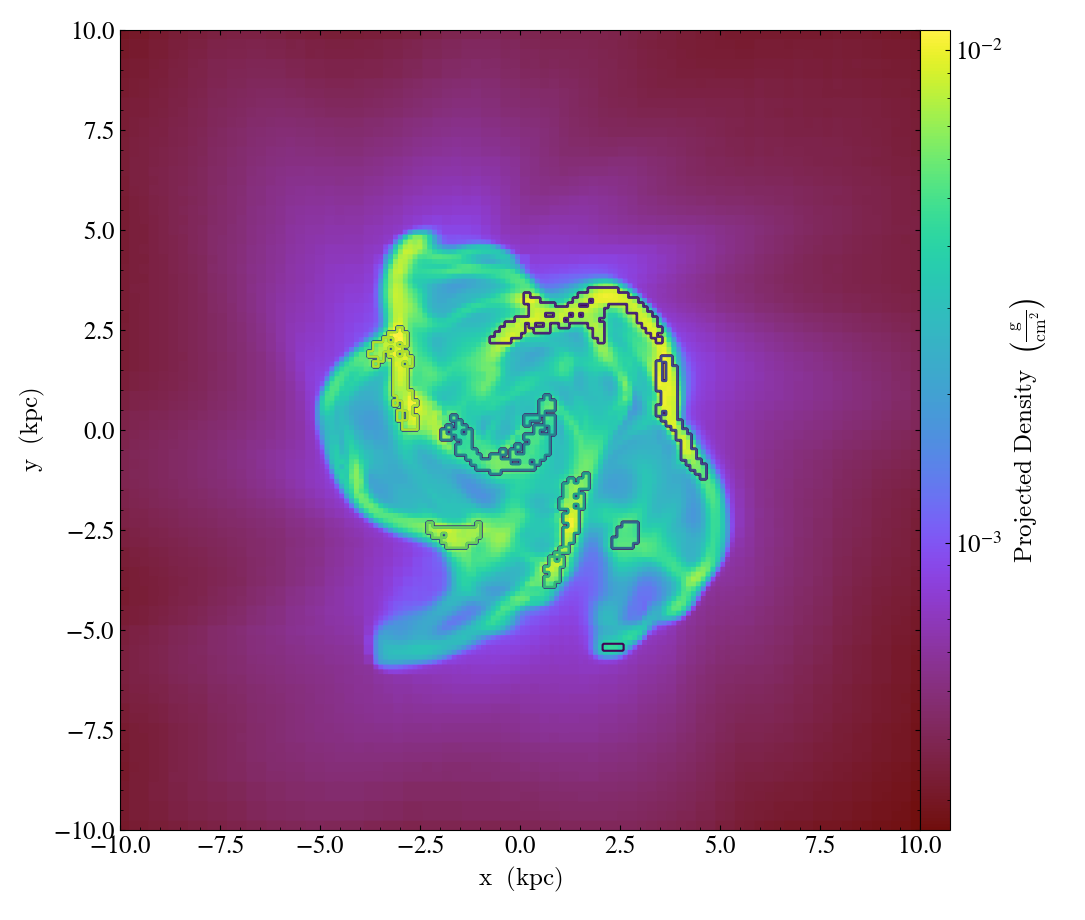
Clump Finder Callback¶
- annotate_clumps(self, clumps, **kwargs)¶
(This is a proxy for
ClumpContourCallback.)Take a list of
clumpsand plot them as a set of contours.
import numpy as np
import yt
from yt.data_objects.level_sets.api import Clump, find_clumps
ds = yt.load("IsolatedGalaxy/galaxy0030/galaxy0030")
data_source = ds.disk([0.5, 0.5, 0.5], [0.0, 0.0, 1.0], (8.0, "kpc"), (1.0, "kpc"))
c_min = 10 ** np.floor(np.log10(data_source["gas", "density"]).min())
c_max = 10 ** np.floor(np.log10(data_source["gas", "density"]).max() + 1)
master_clump = Clump(data_source, ("gas", "density"))
master_clump.add_validator("min_cells", 20)
find_clumps(master_clump, c_min, c_max, 2.0)
leaf_clumps = master_clump.leaves
prj = yt.ProjectionPlot(ds, "z", ("gas", "density"), center="c", width=(20, "kpc"))
prj.annotate_clumps(leaf_clumps)
prj.save("clumps")
Overplot Contours¶
- annotate_contour(self, field, levels=5, factor=4, take_log=False, clim=None, plot_args=None, label=False, text_args=None, data_source=None)¶
(This is a proxy for
ContourCallback.)Add contours in
fieldto the plot.levelsgoverns the number of contours generated,factorgoverns the number of points used in the interpolation,take_loggoverns how it is contoured andclimgives the (lower, upper) limits for contouring.
import yt
ds = yt.load("Enzo_64/DD0043/data0043")
s = yt.SlicePlot(ds, "x", ("gas", "density"), center="max")
s.annotate_contour(("gas", "temperature"))
s.save()
Overplot Quivers¶
Axis-Aligned Data Sources¶
- annotate_quiver(self, field_x, field_y, field_c=None, *, factor=16, scale=None, scale_units=None, normalize=False, **kwargs)¶
(This is a proxy for
QuiverCallback.)Adds a ‘quiver’ plot to any plot, using the
field_xandfield_yfrom the associated data, skipping everyfactorpixels in the discretization. A third field,field_c, can be used as color; which is the counterpart ofmatplotlib.axes.Axes.quiver’s final positional argumentC.scaleis the data units per arrow length unit usingscale_units. IfnormalizeisTrue, the fields will be scaled by their local (in-plane) length, allowing morphological features to be more clearly seen for fields with substantial variation in field strength. All additional keyword arguments are passed down tomatplotlib.Axes.axes.quiver.Example using a constant color
import yt
ds = yt.load("IsolatedGalaxy/galaxy0030/galaxy0030")
p = yt.ProjectionPlot(
ds,
"z",
("gas", "density"),
center=[0.5, 0.5, 0.5],
weight_field="density",
width=(20, "kpc"),
)
p.annotate_quiver(
("gas", "velocity_x"),
("gas", "velocity_y"),
factor=16,
color="purple",
)
p.save()
And now using a continuous colormap
import yt
ds = yt.load("IsolatedGalaxy/galaxy0030/galaxy0030")
p = yt.ProjectionPlot(
ds,
"z",
("gas", "density"),
center=[0.5, 0.5, 0.5],
weight_field="density",
width=(20, "kpc"),
)
p.annotate_quiver(
("gas", "velocity_x"),
("gas", "velocity_y"),
("gas", "vorticity_z"),
factor=16,
cmap="inferno_r",
)
p.save()
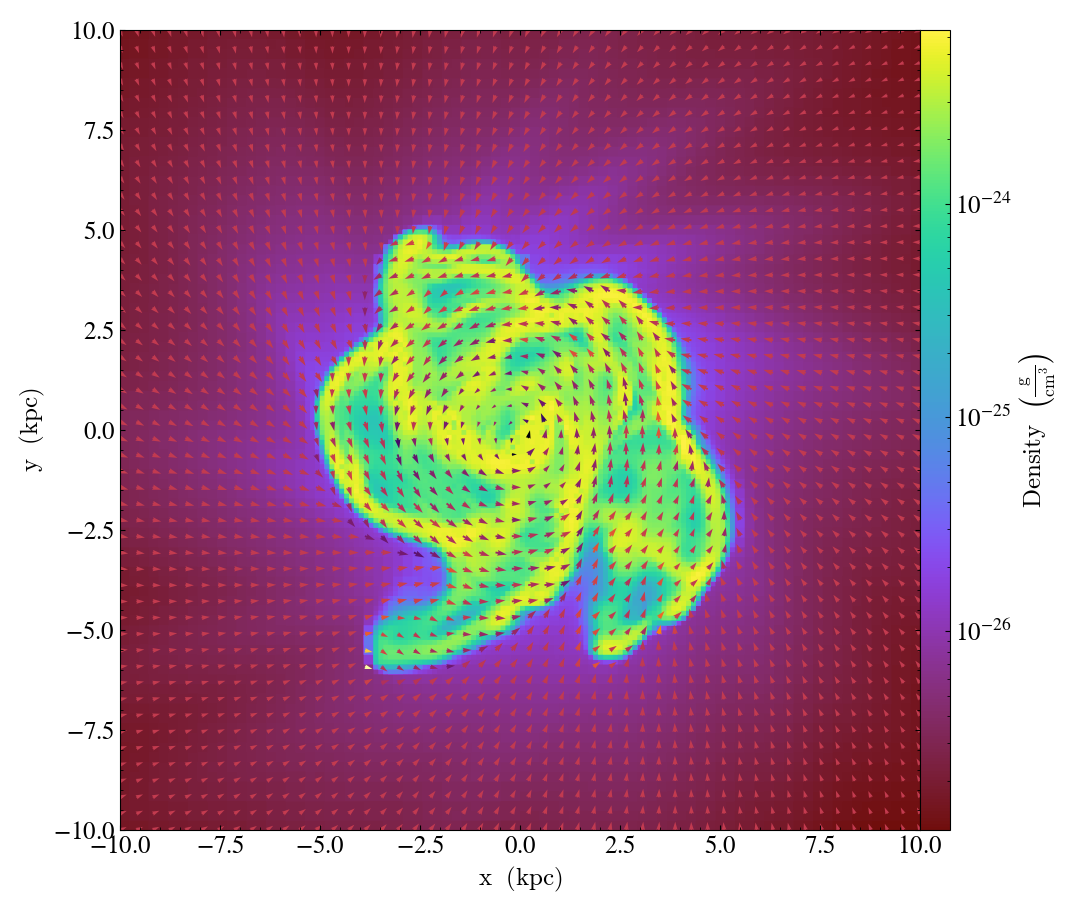
Off-Axis Data Sources¶
- annotate_cquiver(self, field_x, field_y, field_c=None, *, factor=16, scale=None, scale_units=None, normalize=False, **kwargs)¶
(This is a proxy for
CuttingQuiverCallback.)Get a quiver plot on top of a cutting plane, using the
field_xandfield_yfrom the associated data, skipping everyfactordatapoints in the discretization.scaleis the data units per arrow length unit usingscale_units. IfnormalizeisTrue, the fields will be scaled by their local (in-plane) length, allowing morphological features to be more clearly seen for fields with substantial variation in field strength. Additional arguments can be passed to theplot_argsdictionary, see matplotlib.axes.Axes.quiver for more info.
import yt
ds = yt.load("Enzo_64/DD0043/data0043")
s = yt.OffAxisSlicePlot(ds, [1, 1, 0], [("gas", "density")], center="c")
s.annotate_cquiver(
("gas", "cutting_plane_velocity_x"),
("gas", "cutting_plane_velocity_y"),
factor=10,
color="orange",
)
s.zoom(1.5)
s.save()
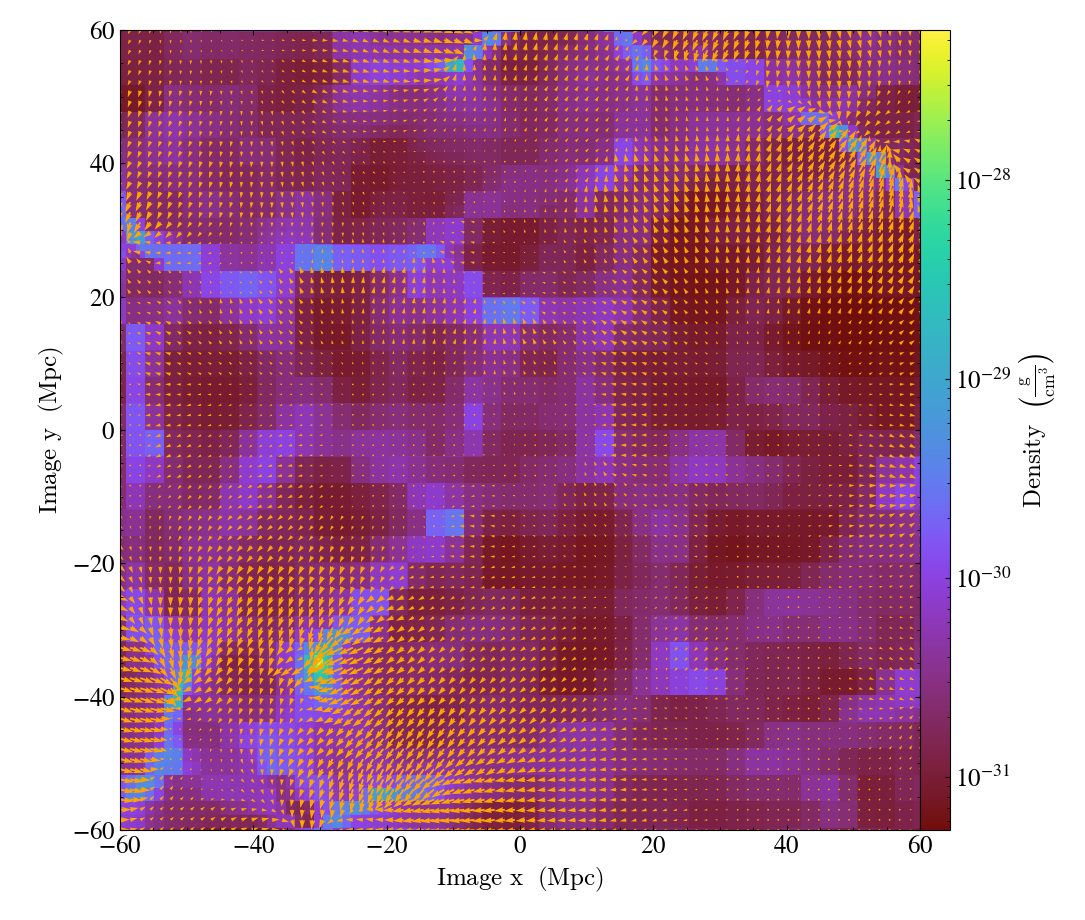
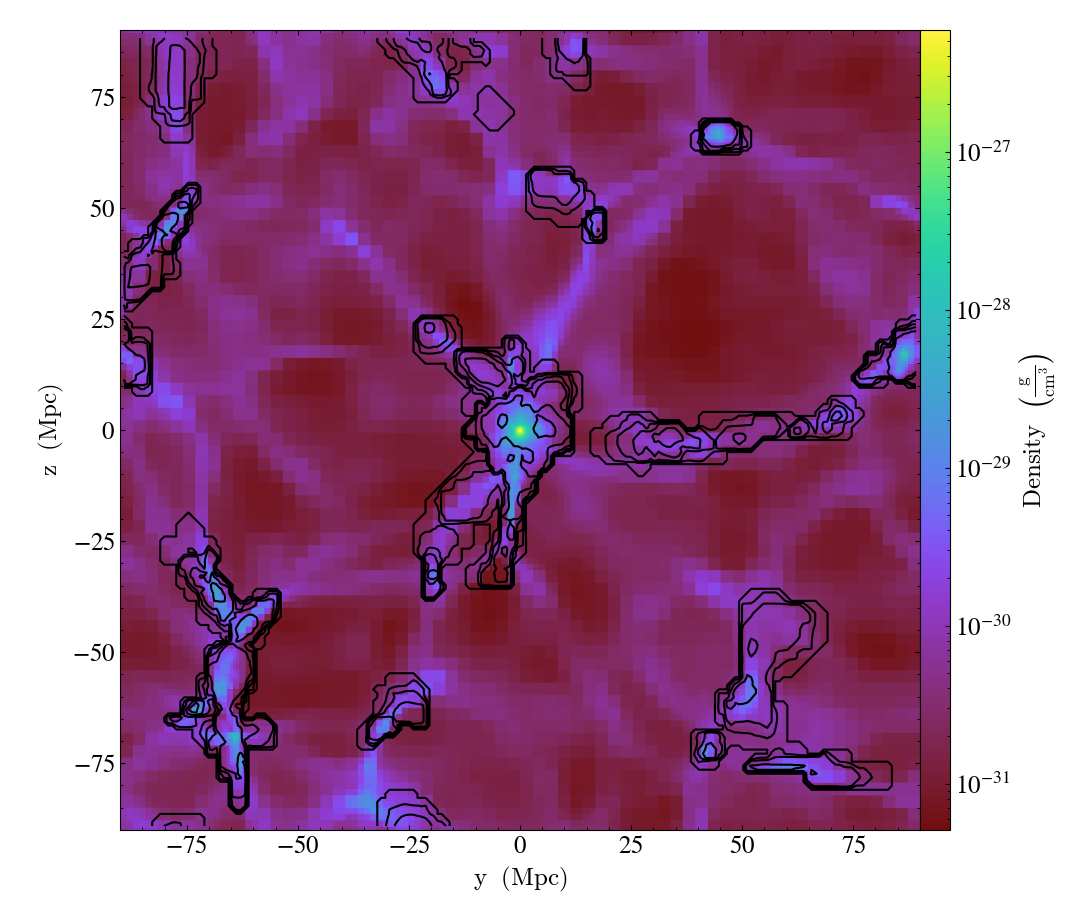
Overplot Grids¶
- annotate_grids(self, alpha=0.7, min_pix=1, min_pix_ids=20, draw_ids=False, id_loc='lower left', periodic=True, min_level=None, max_level=None, cmap='B-W Linear_r', edgecolors=None, linewidth=1.0)¶
(This is a proxy for
GridBoundaryCallback.)Adds grid boundaries to a plot, optionally with alpha-blending via the
alphakeyword. Cuttoff for display is atmin_pixwide.draw_idsputs the grid id in theid_loccorner of the grid. (id_loccan be upper/lower left/right.draw_idsis not so great in projections…)
import yt
ds = yt.load("IsolatedGalaxy/galaxy0030/galaxy0030")
slc = yt.SlicePlot(ds, "z", ("gas", "density"), width=(10, "kpc"), center="max")
slc.annotate_grids()
slc.save()
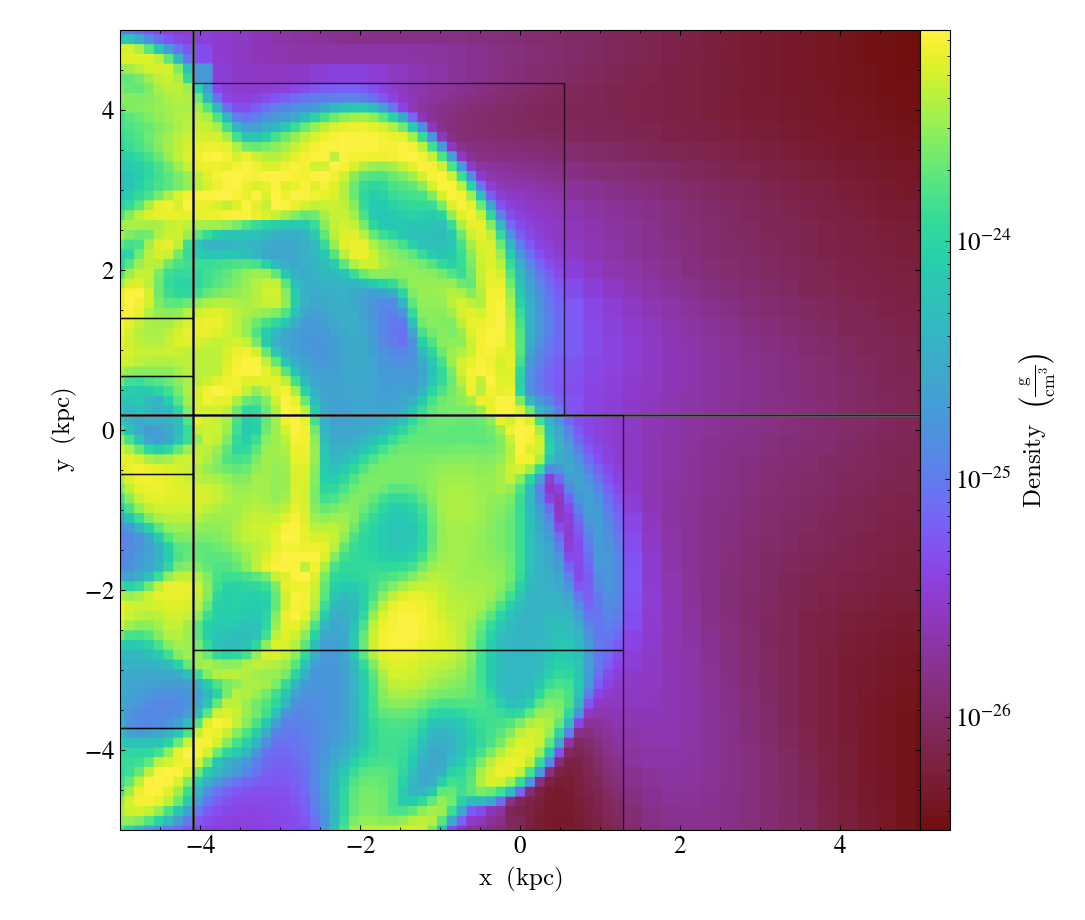
Overplot Cell Edges¶
- annotate_cell_edges(line_width=0.002, alpha=1.0, color='black')¶
(This is a proxy for
CellEdgesCallback.)Annotate the edges of cells, where the
line_widthrelative to size of the longest plot axis is specified. Thealphaof the overlaid image and thecolorof the lines are also specifiable. Note that because the lines are drawn from both sides of a cell, the image sometimes has the effect of doubling the line width. Color here is a matplotlib color name or a 3-tuple of RGB float values.
import yt
ds = yt.load("IsolatedGalaxy/galaxy0030/galaxy0030")
slc = yt.SlicePlot(ds, "z", ("gas", "density"), width=(10, "kpc"), center="max")
slc.annotate_cell_edges()
slc.save()
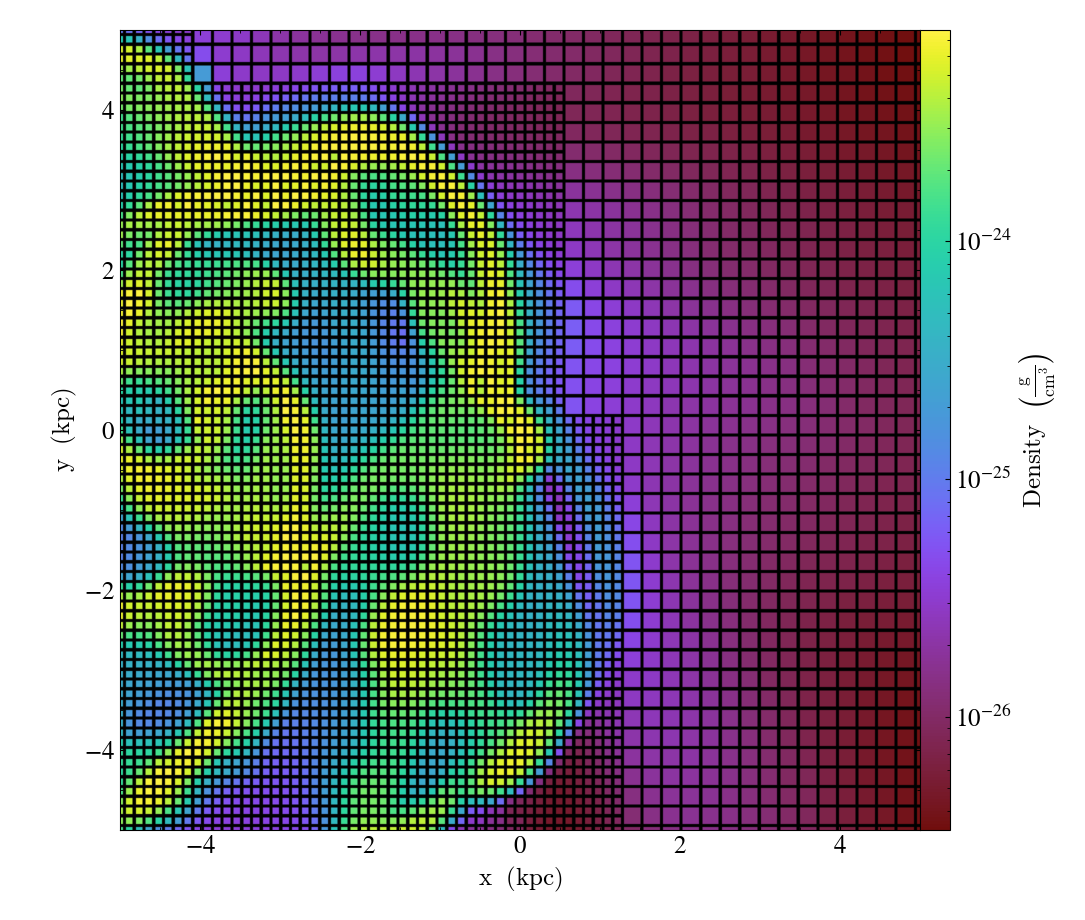
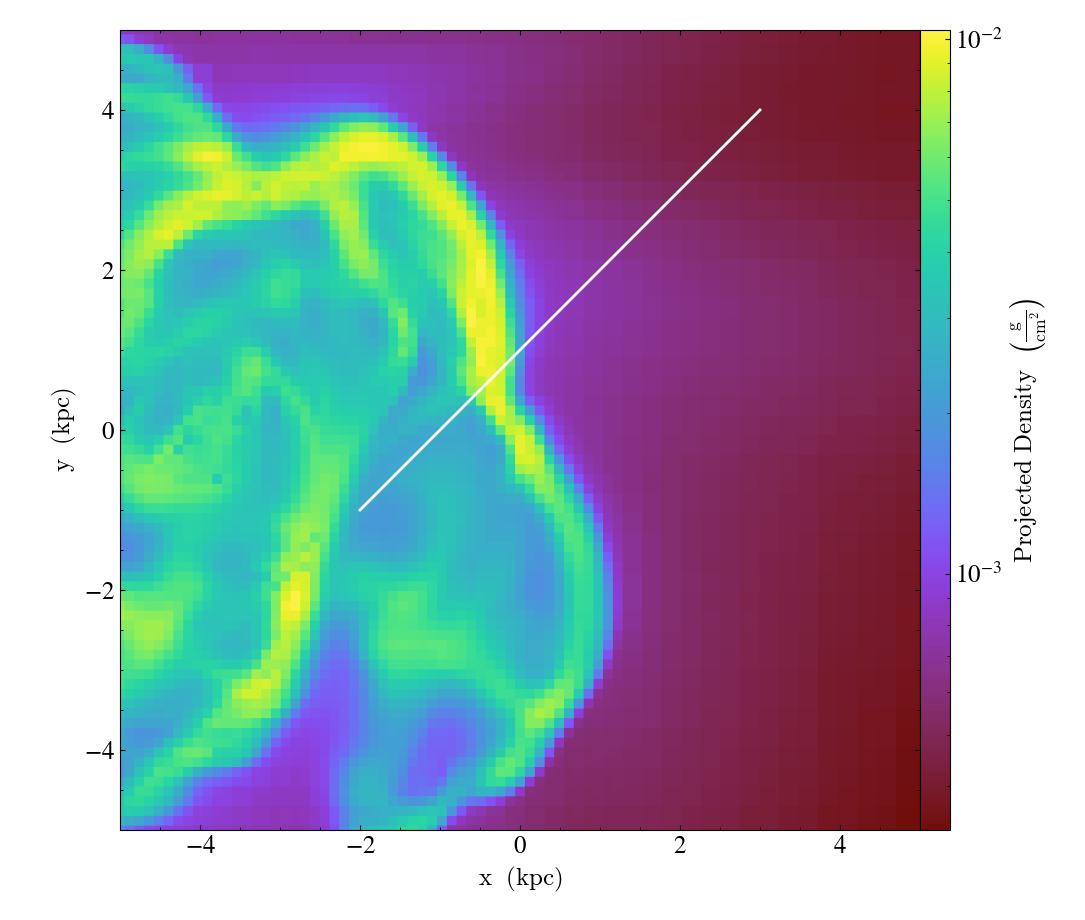
Overplot a Straight Line¶
- annotate_line(self, p1, p2, *, coord_system='data', **kwargs)¶
(This is a proxy for
LinePlotCallback.)Overplot a line with endpoints at p1 and p2. p1 and p2 should be 2D or 3D coordinates consistent with the coordinate system denoted in the “coord_system” keyword.
import yt
ds = yt.load("IsolatedGalaxy/galaxy0030/galaxy0030")
p = yt.ProjectionPlot(ds, "z", ("gas", "density"), center="m", width=(10, "kpc"))
p.annotate_line((0.3, 0.4), (0.8, 0.9), coord_system="axis")
p.save()
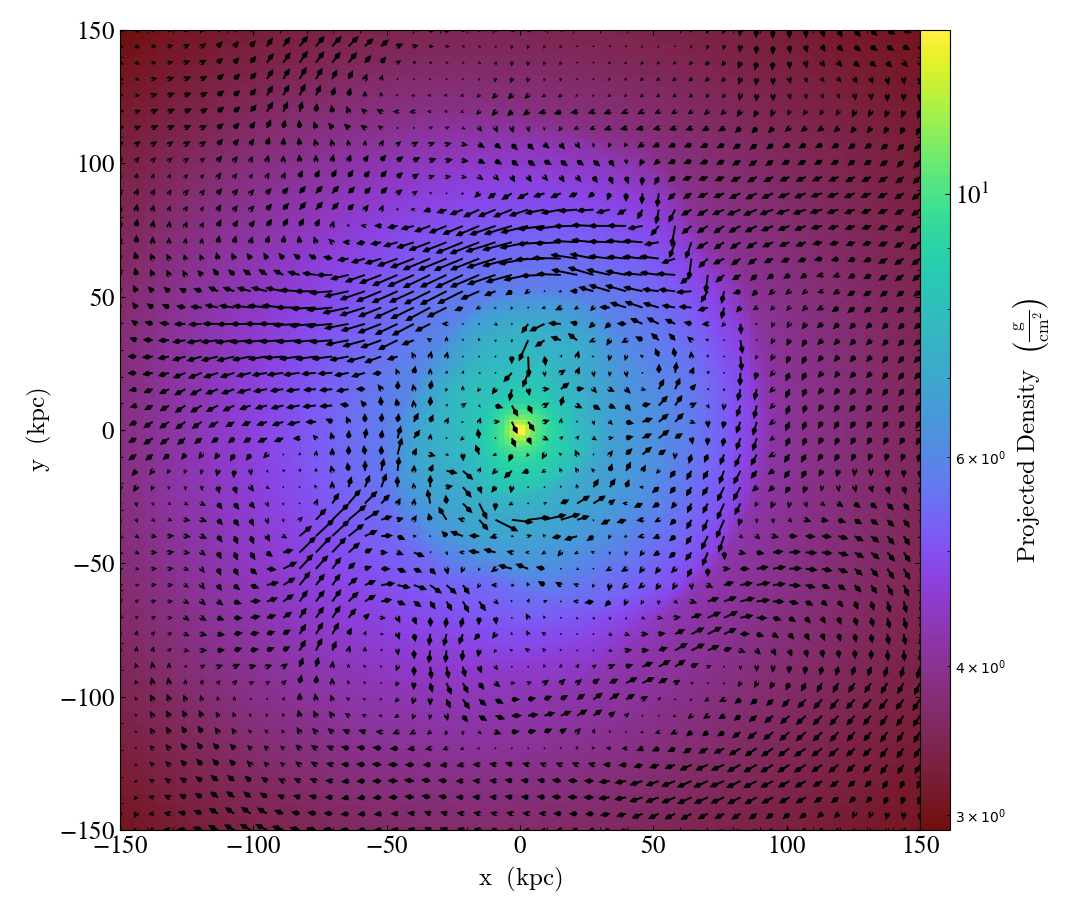
Overplot Magnetic Field Quivers¶
- annotate_magnetic_field(self, factor=16, *, scale=None, scale_units=None, normalize=False, **kwargs)¶
(This is a proxy for
MagFieldCallback.)Adds a ‘quiver’ plot of magnetic field to the plot, skipping every
factordatapoints in the discretization.scaleis the data units per arrow length unit usingscale_units. IfnormalizeisTrue, the magnetic fields will be scaled by their local (in-plane) length, allowing morphological features to be more clearly seen for fields with substantial variation in field strength. Additional arguments can be passed to theplot_argsdictionary, see matplotlib.axes.Axes.quiver for more info.
import yt
ds = yt.load(
"MHDSloshing/virgo_low_res.0054.vtk",
units_override={
"time_unit": (1, "Myr"),
"length_unit": (1, "Mpc"),
"mass_unit": (1e17, "Msun"),
},
)
p = yt.ProjectionPlot(ds, "z", ("gas", "density"), center="c", width=(300, "kpc"))
p.annotate_magnetic_field(headlength=3)
p.save()
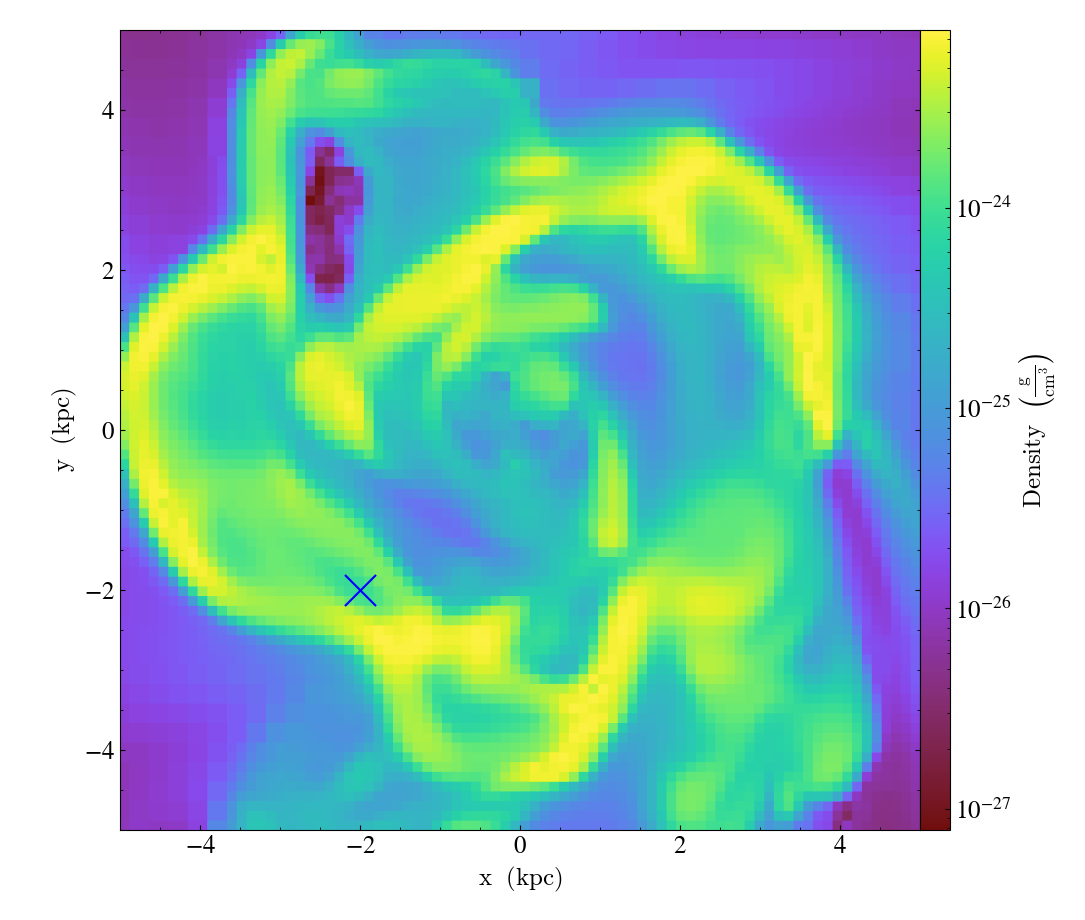
Annotate a Point With a Marker¶
- annotate_marker(self, pos, marker='x', *, coord_system='data', **kwargs)¶
(This is a proxy for
MarkerAnnotateCallback.)Overplot a marker on a position for highlighting specific features.
import yt
ds = yt.load("IsolatedGalaxy/galaxy0030/galaxy0030")
s = yt.SlicePlot(ds, "z", ("gas", "density"), center="c", width=(10, "kpc"))
s.annotate_marker((-2, -2), coord_system="plot", color="blue", s=500)
s.save()
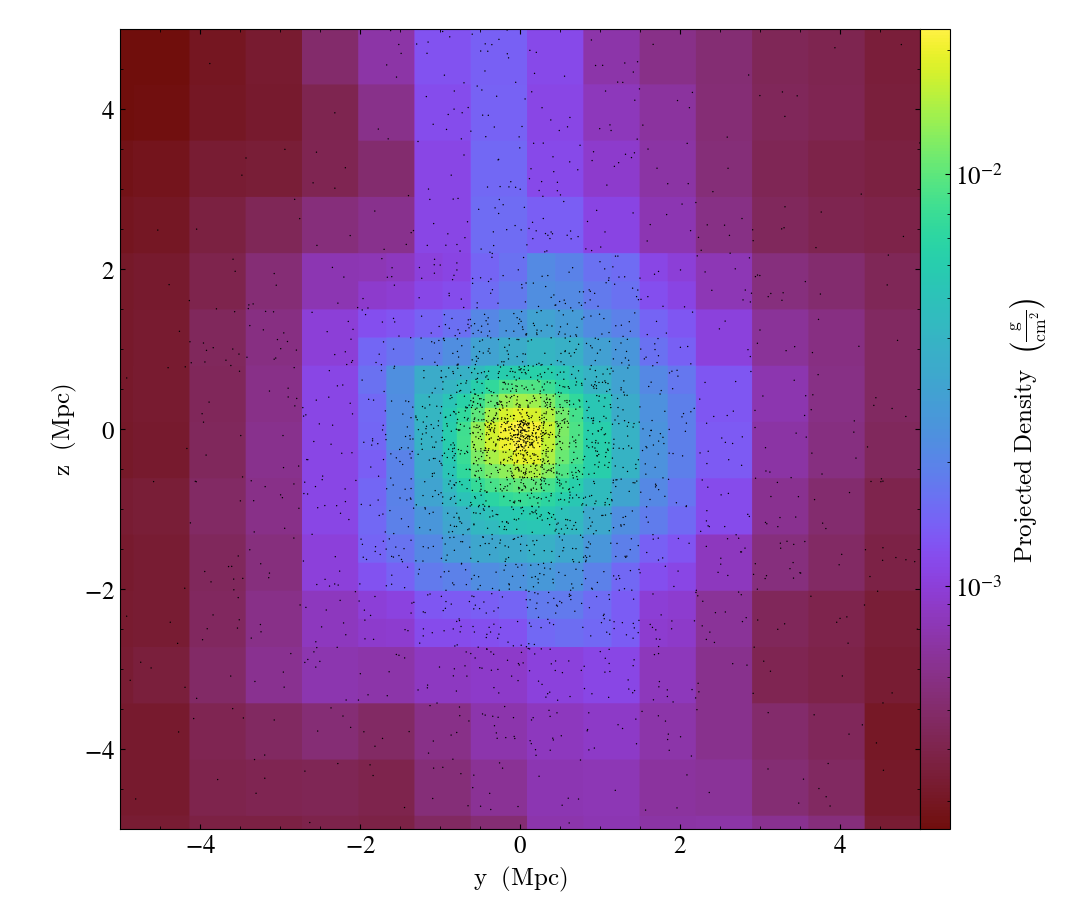
Overplotting Particle Positions¶
- annotate_particles(self, width, p_size=1.0, col='k', marker='o', stride=1, ptype='all', alpha=1.0, data_source=None)¶
(This is a proxy for
ParticleCallback.)Adds particle positions, based on a thick slab along
axiswith awidthalong the line of sight.p_sizecontrols the number of pixels per particle, andcolgoverns the color.ptypewill restrict plotted particles to only those that are of a given type.data_sourcewill only plot particles contained within the data_source object.WARNING: if
data_sourceis ayt.data_objects.selection_data_containers.YTCutRegionthen thewidthparameter is ignored.
import yt
ds = yt.load("Enzo_64/DD0043/data0043")
p = yt.ProjectionPlot(ds, "x", ("gas", "density"), center="m", width=(10, "Mpc"))
p.annotate_particles((10, "Mpc"))
p.save()
To plot only the central particles
import yt
ds = yt.load("Enzo_64/DD0043/data0043")
p = yt.ProjectionPlot(ds, "x", ("gas", "density"), center="m", width=(10, "Mpc"))
sp = ds.sphere(p.data_source.center, ds.quan(1, "Mpc"))
p.annotate_particles((10, "Mpc"), data_source=sp)
p.save()
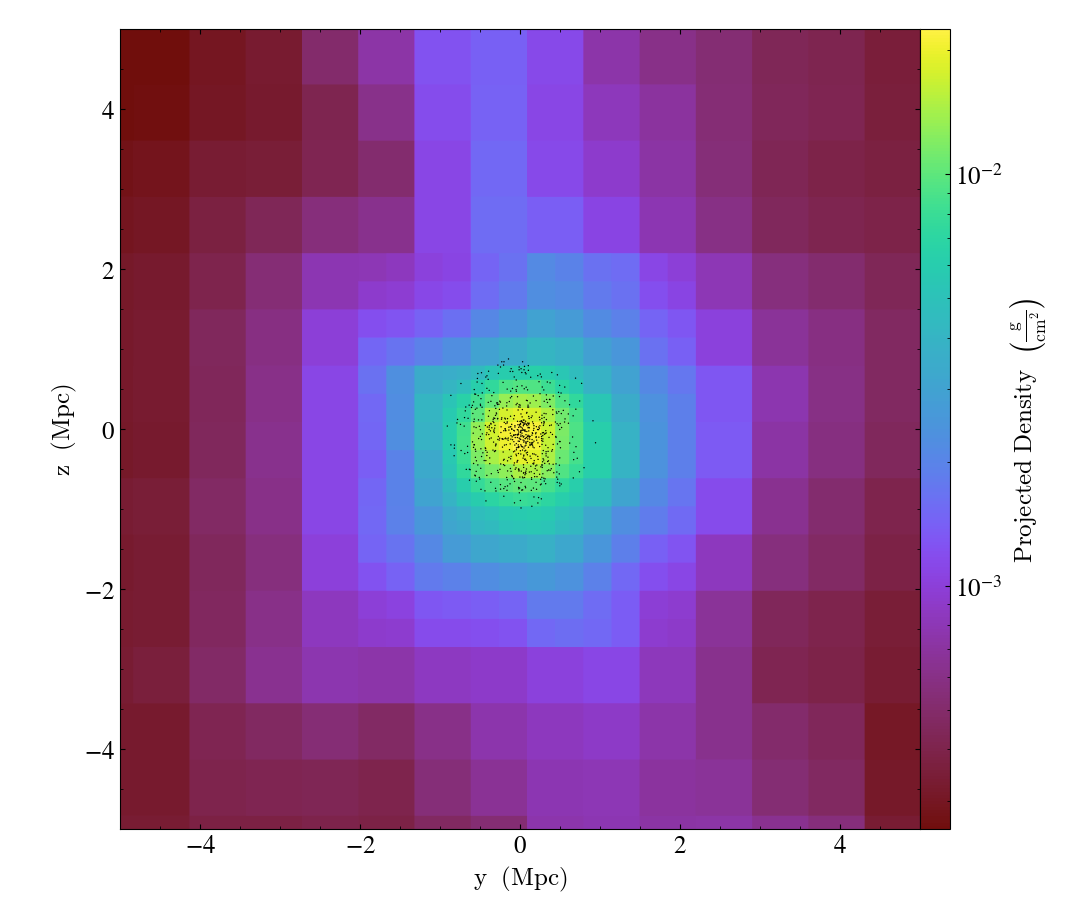
Overplot a Circle on a Plot¶
- annotate_sphere(self, center, radius, circle_args=None, coord_system='data', text=None, text_args=None)¶
(This is a proxy for
SphereCallback.)Overplot a circle with designated center and radius with optional text.
import yt
ds = yt.load("IsolatedGalaxy/galaxy0030/galaxy0030")
p = yt.ProjectionPlot(ds, "z", ("gas", "density"), center="c", width=(20, "kpc"))
p.annotate_sphere([0.5, 0.5, 0.5], radius=(2, "kpc"), circle_args={"color": "black"})
p.save()
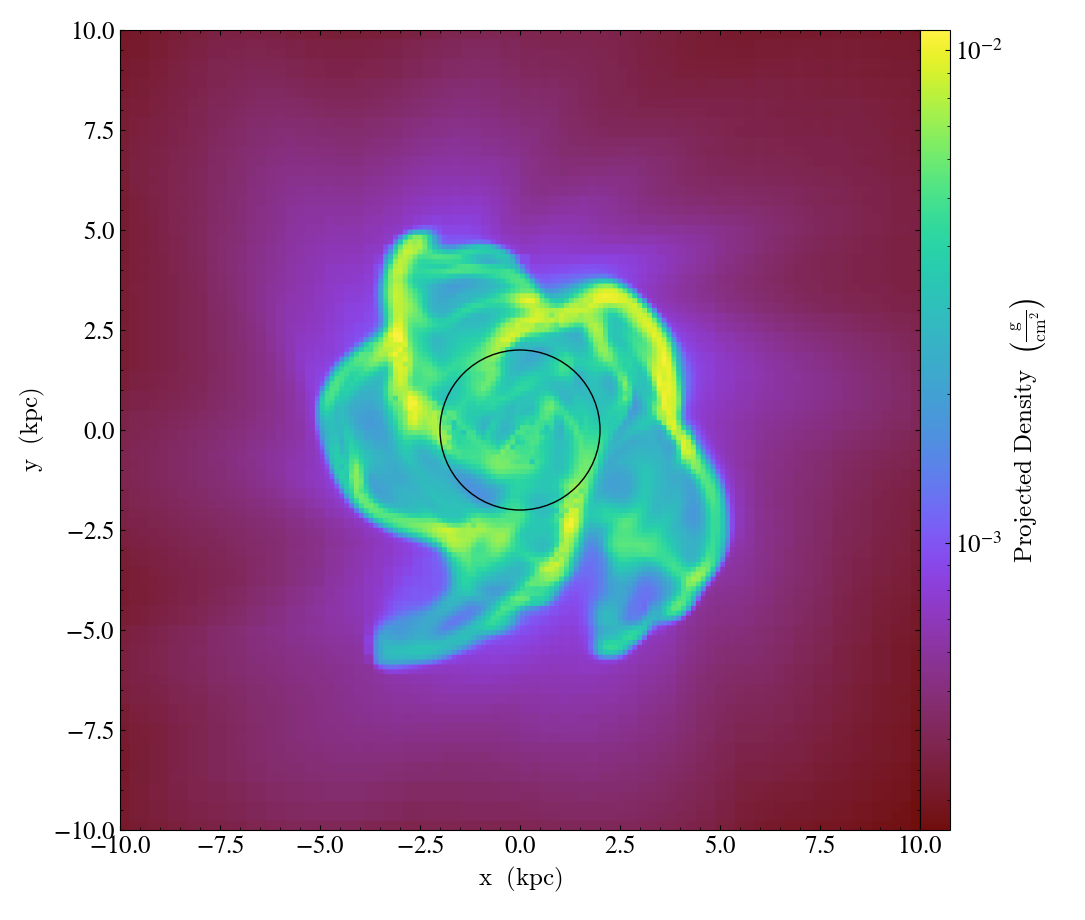
Overplot Streamlines¶
- annotate_streamlines(self, field_x, field_y, *, linewidth=1.0, linewidth_upscaling=1.0, color=None, color_threshold=float('-inf'), factor=16, **kwargs)¶
(This is a proxy for
StreamlineCallback.)Add streamlines to any plot, using the
field_xandfield_yfrom the associated data, usingnxandnystarting points that are bounded byxstartandystart. To begin streamlines from the left edge of the plot, setstart_at_xedgetoTrue; for the bottom edge, usestart_at_yedge. A line with the qmean vector magnitude will cover 1.0/factorof the image.Additional keyword arguments are passed down to matplotlib.axes.Axes.streamplot
import yt
ds = yt.load("IsolatedGalaxy/galaxy0030/galaxy0030")
s = yt.SlicePlot(ds, "z", ("gas", "density"), center="c", width=(20, "kpc"))
s.annotate_streamlines(("gas", "velocity_x"), ("gas", "velocity_y"))
s.save()
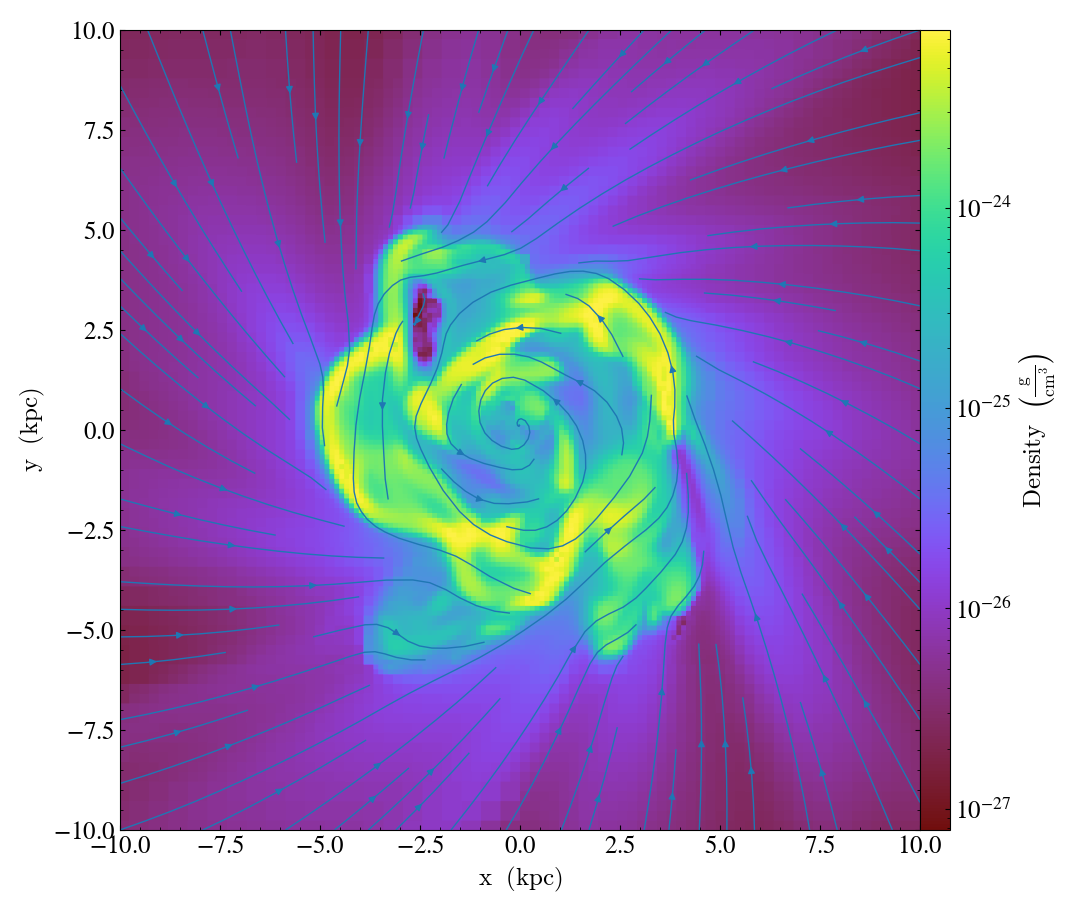
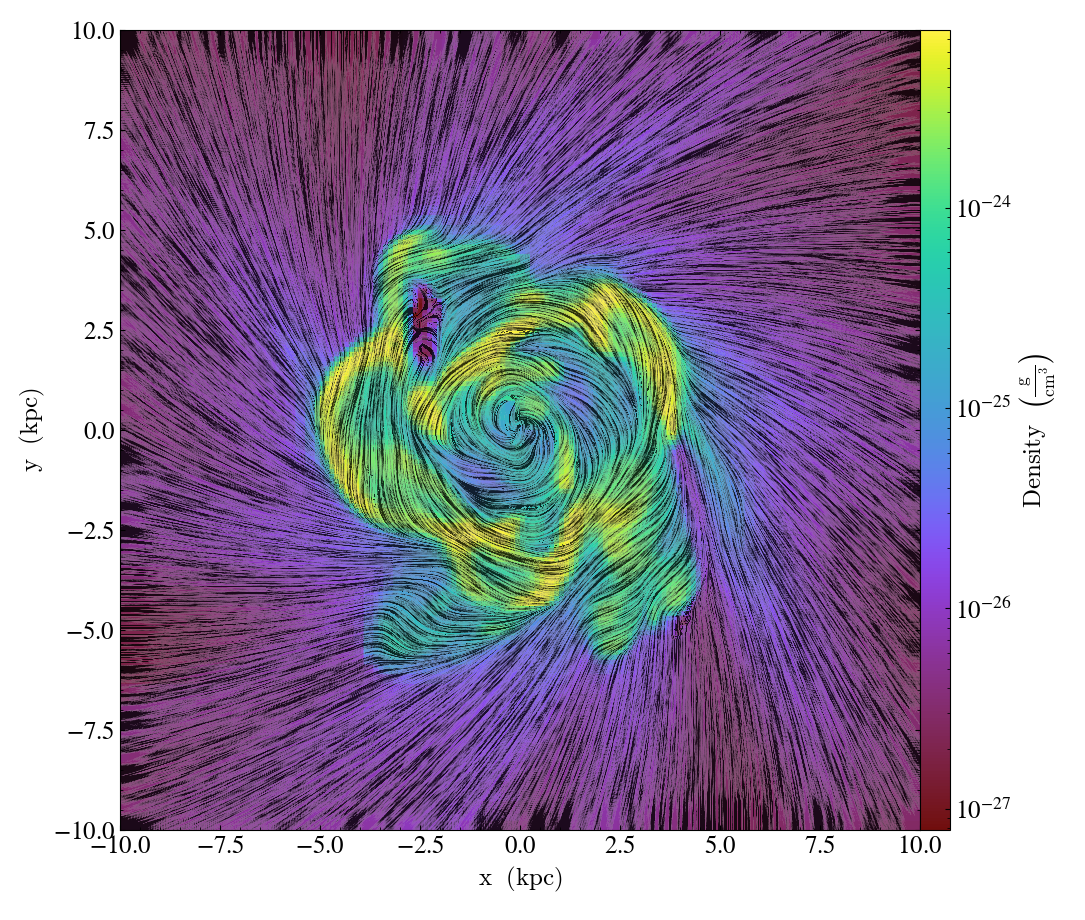
Overplot Line Integral Convolution¶
- annotate_line_integral_convolution(self, field_x, field_y, texture=None, kernellen=50., lim=(0.5, 0.6), cmap='binary', alpha=0.8, const_alpha=False)¶
(This is a proxy for
LineIntegralConvolutionCallback.)Add line integral convolution to any plot, using the
field_xandfield_yfrom the associated data. A white noise background will be used fortextureas default. Adjust the bounds oflimin the range of[0, 1]which applies upper and lower bounds to the values of line integral convolution and enhance the visibility of plots. Whenconst_alpha=False, alpha will be weighted spatially by the values of line integral convolution; otherwise a constant value of the given alpha is used.
import yt
ds = yt.load("IsolatedGalaxy/galaxy0030/galaxy0030")
s = yt.SlicePlot(ds, "z", ("gas", "density"), center="c", width=(20, "kpc"))
s.annotate_line_integral_convolution(("gas", "velocity_x"), ("gas", "velocity_y"), lim=(0.5, 0.65))
s.save()
Overplot Text¶
- annotate_text(self, pos, text, coord_system='data', text_args=None, inset_box_args=None)¶
(This is a proxy for
TextLabelCallback.)Overplot text on the plot at a specified position. If you desire an inset box around your text, set one with the inset_box_args dictionary keyword.
import yt
ds = yt.load("IsolatedGalaxy/galaxy0030/galaxy0030")
s = yt.SlicePlot(ds, "z", ("gas", "density"), center="max", width=(10, "kpc"))
s.annotate_text((2, 2), "Galaxy!", coord_system="plot")
s.save()
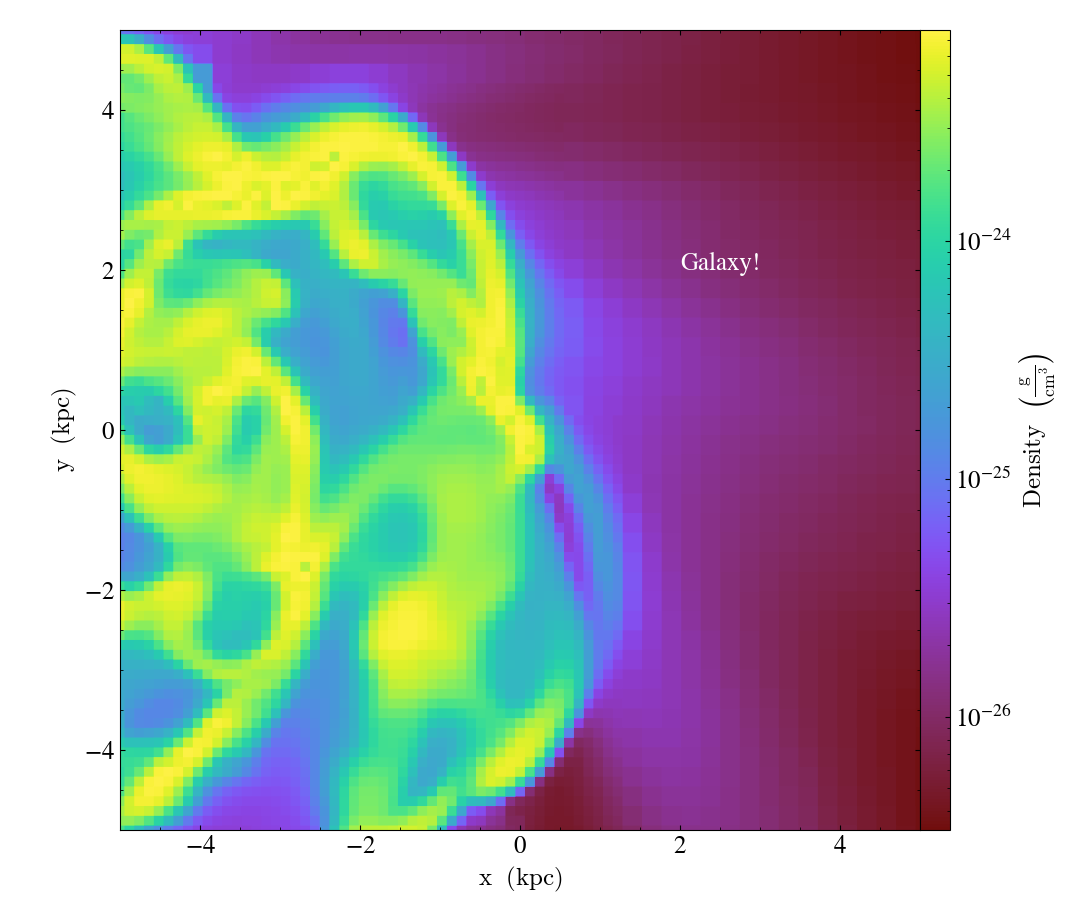
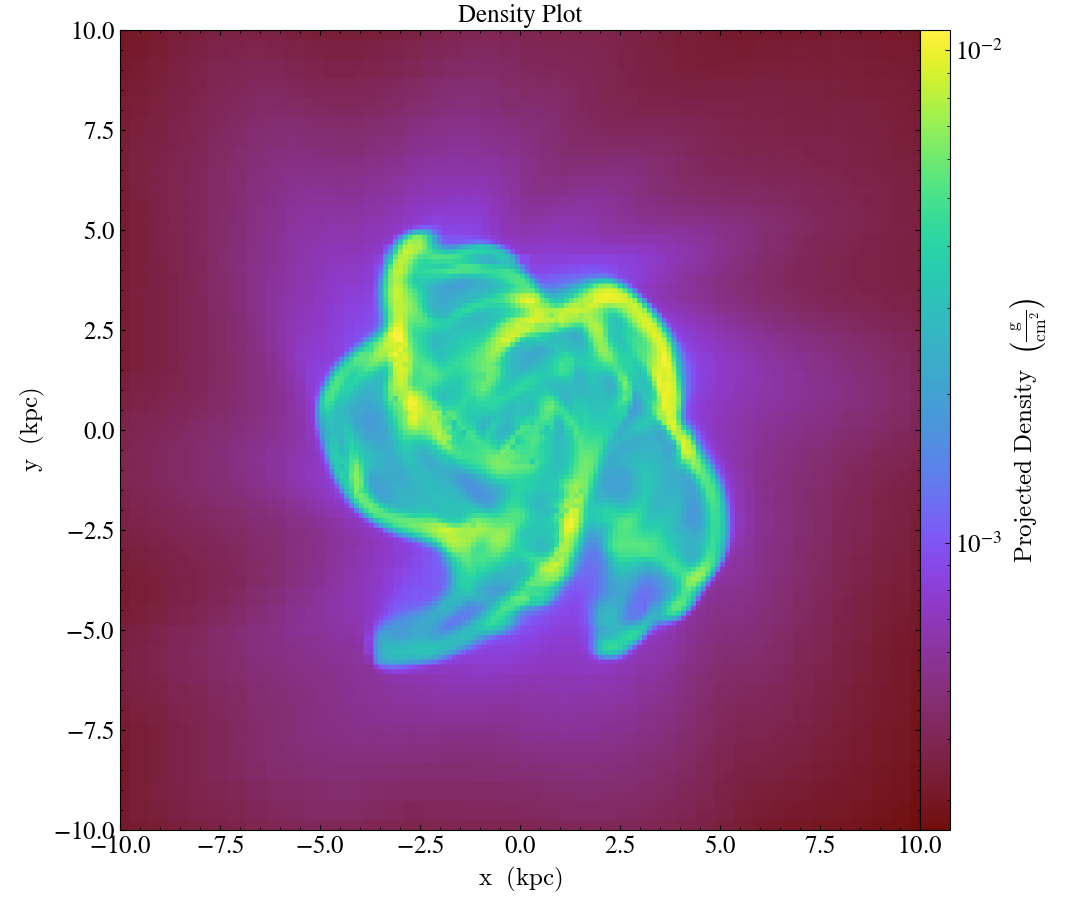
Add a Title¶
- annotate_title(self, title='Plot')¶
(This is a proxy for
TitleCallback.)Accepts a
titleand adds it to the plot.
import yt
ds = yt.load("IsolatedGalaxy/galaxy0030/galaxy0030")
p = yt.ProjectionPlot(ds, "z", ("gas", "density"), center="c", width=(20, "kpc"))
p.annotate_title("Density Plot")
p.save()
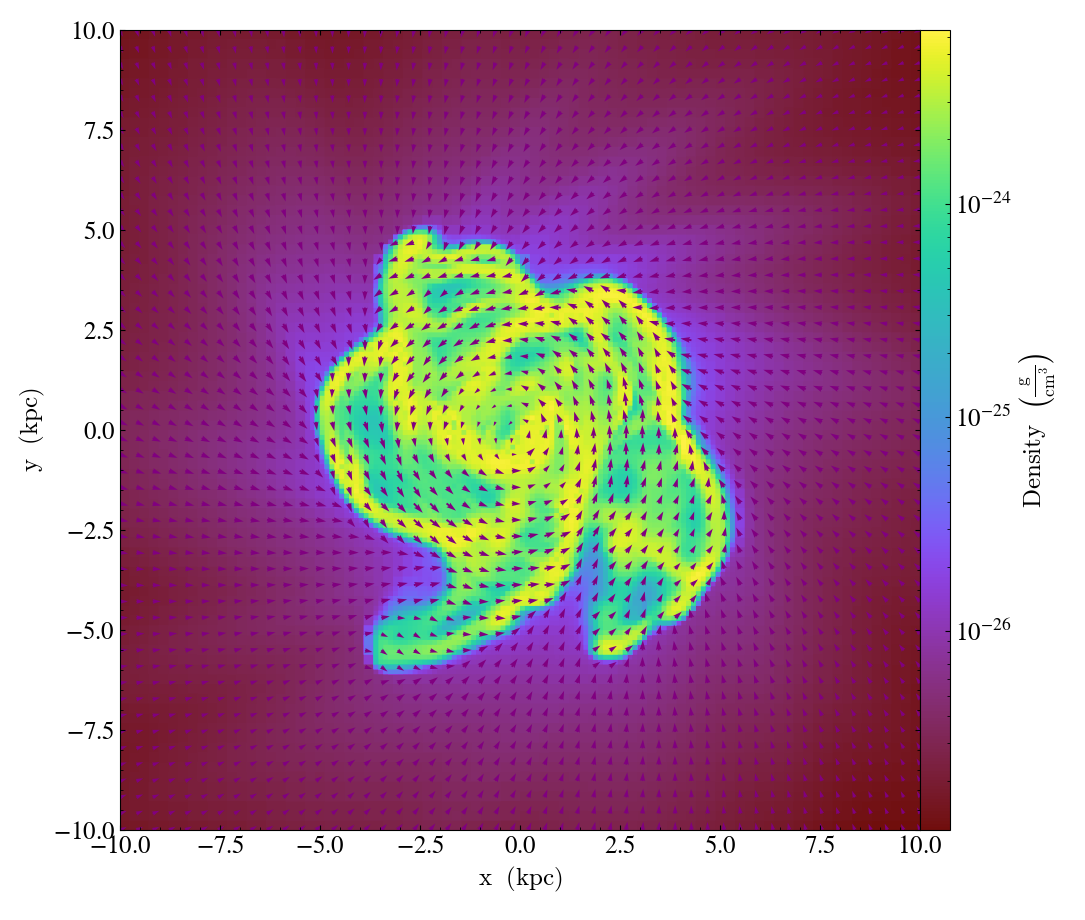
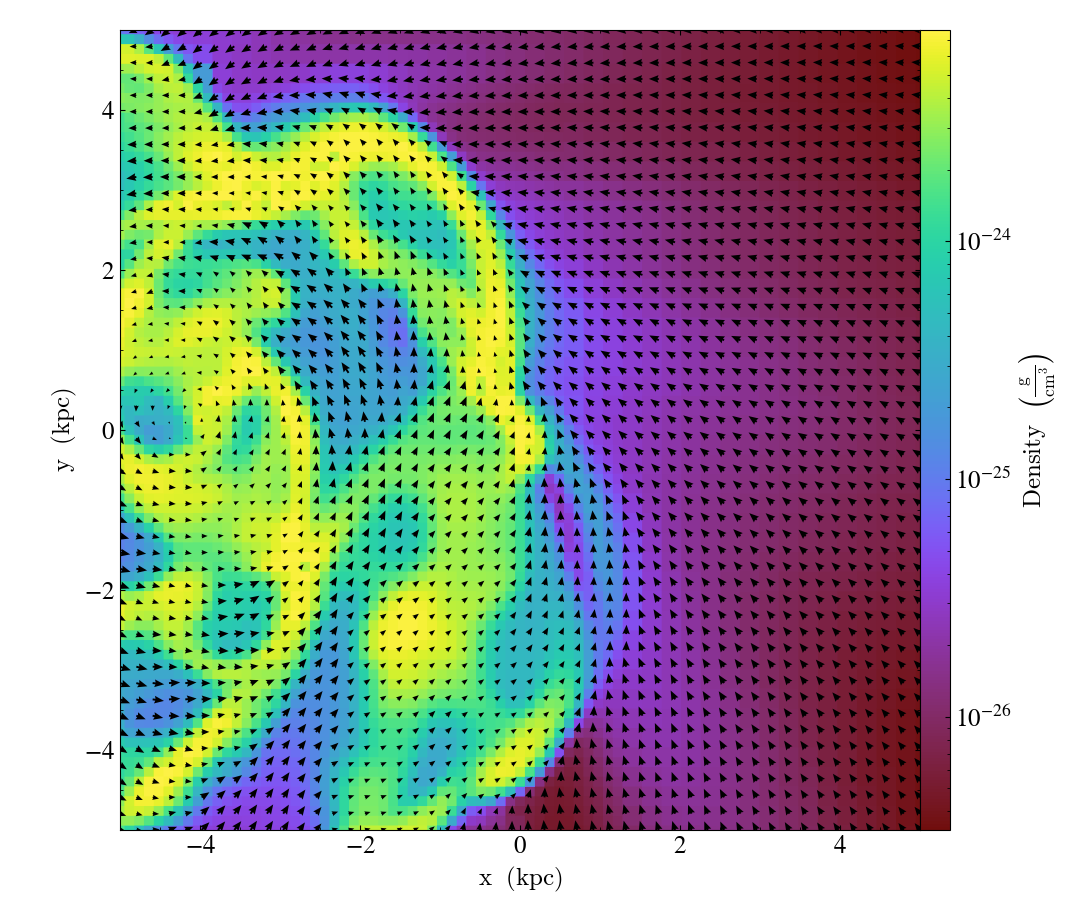
Overplot Quivers for the Velocity Field¶
- annotate_velocity(self, factor=16, *, scale=None, scale_units=None, normalize=False, **kwargs)¶
(This is a proxy for
VelocityCallback.)Adds a ‘quiver’ plot of velocity to the plot, skipping every
factordatapoints in the discretization.scaleis the data units per arrow length unit usingscale_units. IfnormalizeisTrue, the velocity fields will be scaled by their local (in-plane) length, allowing morphological features to be more clearly seen for fields with substantial variation in field strength. Additional arguments can be passed to theplot_argsdictionary, see matplotlib.axes.Axes.quiver for more info.
import yt
ds = yt.load("IsolatedGalaxy/galaxy0030/galaxy0030")
p = yt.SlicePlot(ds, "z", ("gas", "density"), center="m", width=(10, "kpc"))
p.annotate_velocity(headwidth=4)
p.save()
Add the Current Time and/or Redshift¶
- annotate_timestamp(x_pos=None, y_pos=None, corner='lower_left', time=True, redshift=False, time_format='t = {time:.1f} {units}', time_unit=None, time_offset=None, redshift_format='z = {redshift:.2f}', draw_inset_box=False, coord_system='axis', text_args=None, inset_box_args=None)¶
(This is a proxy for
TimestampCallback.)Annotates the timestamp and/or redshift of the data output at a specified location in the image (either in a present corner, or by specifying (x,y) image coordinates with the x_pos, y_pos arguments. If no time_units are specified, it will automatically choose appropriate units. It allows for custom formatting of the time and redshift information, the specification of an inset box around the text, and changing the value of the timestamp via a constant offset.
import yt
ds = yt.load("IsolatedGalaxy/galaxy0030/galaxy0030")
p = yt.SlicePlot(ds, "z", ("gas", "density"), center="c", width=(20, "kpc"))
p.annotate_timestamp()
p.save()
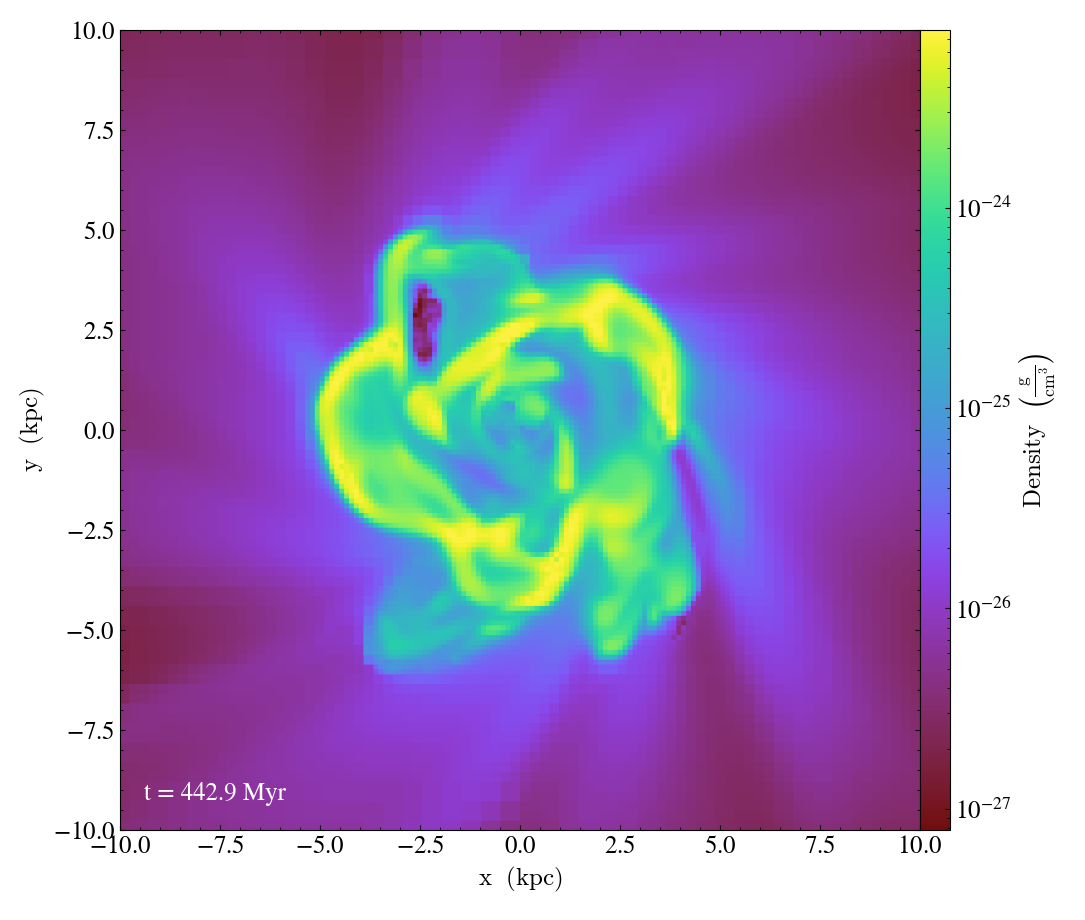
Add a Physical Scale Bar¶
- annotate_scale(corner='lower_right', coeff=None, unit=None, pos=None, scale_text_format='{scale} {units}', max_frac=0.16, min_frac=0.015, coord_system='axis', text_args=None, size_bar_args=None, draw_inset_box=False, inset_box_args=None)¶
(This is a proxy for
ScaleCallback.)Annotates the scale of the plot at a specified location in the image (either in a preset corner, or by specifying (x,y) image coordinates with the pos argument. Coeff and units (e.g. 1 Mpc or 100 kpc) refer to the distance scale you desire to show on the plot. If no coeff and units are specified, an appropriate pair will be determined such that your scale bar is never smaller than min_frac or greater than max_frac of your plottable axis length. Additional customization of the scale bar is possible by adjusting the text_args and size_bar_args dictionaries. The text_args dictionary accepts matplotlib’s font_properties arguments to override the default font_properties for the current plot. The size_bar_args dictionary accepts keyword arguments for the AnchoredSizeBar class in matplotlib’s axes_grid toolkit. Finally, the format of the scale bar text can be adjusted using the scale_text_format keyword argument.
import yt
ds = yt.load("IsolatedGalaxy/galaxy0030/galaxy0030")
p = yt.SlicePlot(ds, "z", ("gas", "density"), center="c", width=(20, "kpc"))
p.annotate_scale()
p.save()
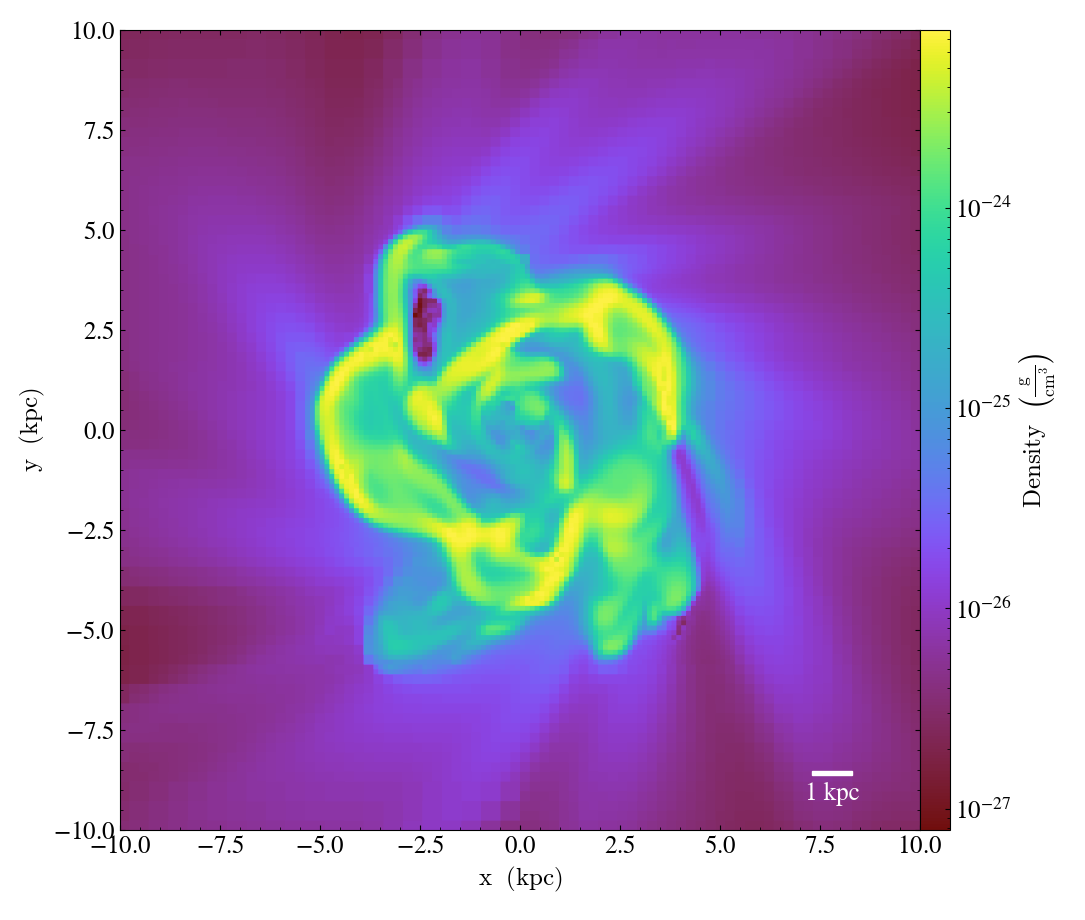
Annotate Triangle Facets Callback¶
- annotate_triangle_facets(triangle_vertices, **kwargs)¶
(This is a proxy for
TriangleFacetsCallback.)This add a line collection of a SlicePlot’s plane-intersection with the triangles to the plot. This callback is ideal for a dataset representing a geometric model of triangular facets.
import os
import h5py
import yt
# Load data file
ds = yt.load("MoabTest/fng_usrbin22.h5m")
# Create the desired slice plot
s = yt.SlicePlot(ds, "z", ("moab", "TALLY_TAG"))
# get triangle vertices from file (in this case hdf5)
# setup file path for yt test directory
filename = os.path.join(
yt.config.ytcfg.get("yt", "test_data_dir"), "MoabTest/mcnp_n_impr_fluka.h5m"
)
f = h5py.File(filename, mode="r")
coords = f["/tstt/nodes/coordinates"][:]
conn = f["/tstt/elements/Tri3/connectivity"][:]
points = coords[conn - 1]
# Annotate slice-triangle intersection contours to the plot
s.annotate_triangle_facets(points, colors="black")
s.save()

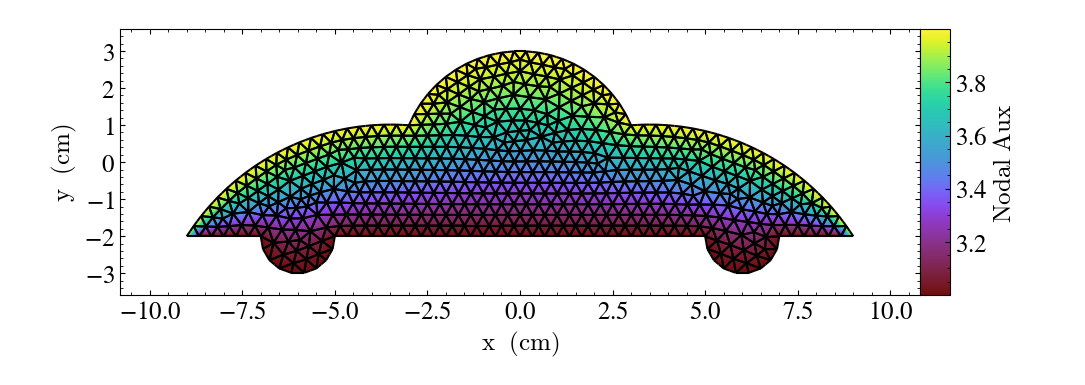
Annotate Mesh Lines Callback¶
- annotate_mesh_lines(**kwargs)¶
(This is a proxy for
MeshLinesCallback.)This draws the mesh line boundaries over a plot using a Matplotlib line collection. This callback is only useful for unstructured or semi-structured mesh datasets.
import yt
ds = yt.load("MOOSE_sample_data/out.e")
sl = yt.SlicePlot(ds, "z", ("connect1", "nodal_aux"))
sl.annotate_mesh_lines(color="black")
sl.save()
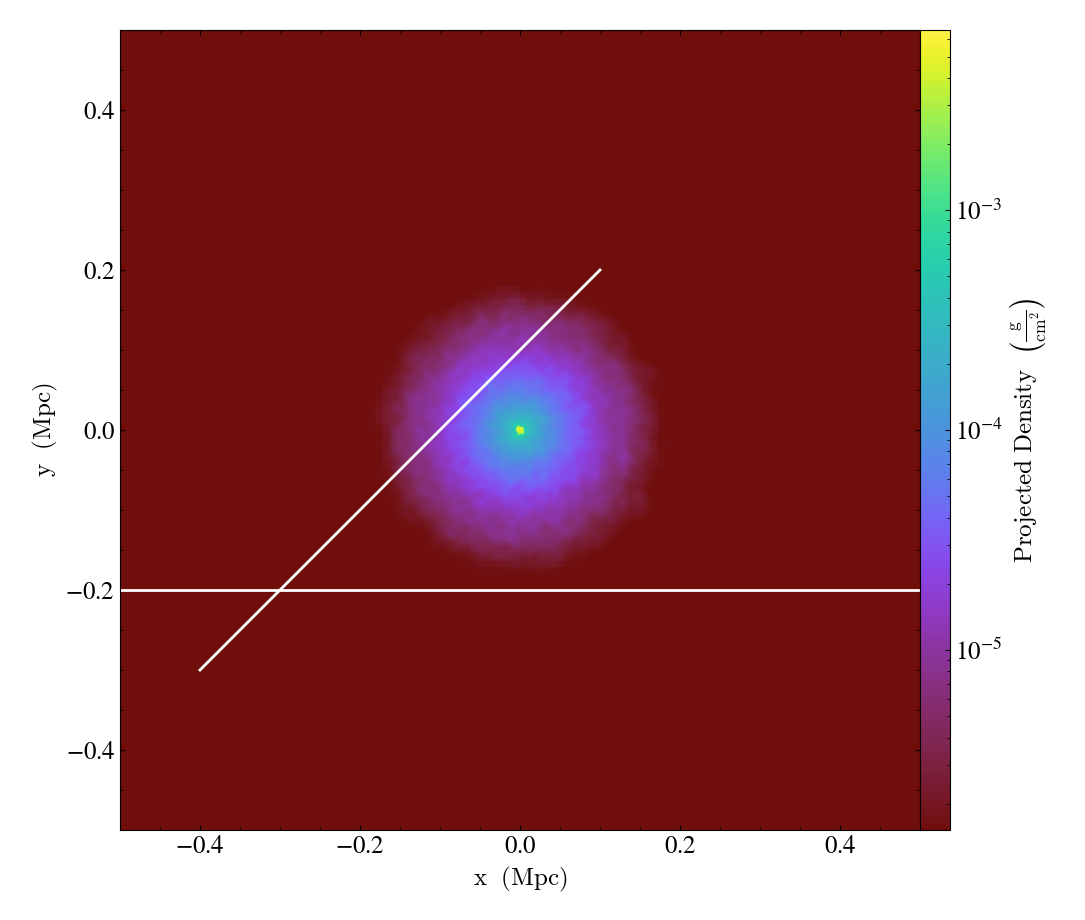
Overplot the Path of a Ray¶
- annotate_ray(ray, *, arrow=False, **kwargs)¶
(This is a proxy for
RayCallback.)Adds a line representing the projected path of a ray across the plot. The ray can be either a
YTOrthoRay,YTRay, or a TridentLightRayobject. annotate_ray() will properly account for periodic rays across the volume.
import yt
ds = yt.load("IsolatedGalaxy/galaxy0030/galaxy0030")
oray = ds.ortho_ray(0, (0.3, 0.4))
ray = ds.ray((0.1, 0.2, 0.3), (0.6, 0.7, 0.8))
p = yt.ProjectionPlot(ds, "z", ("gas", "density"))
p.annotate_ray(oray)
p.annotate_ray(ray)
p.save()
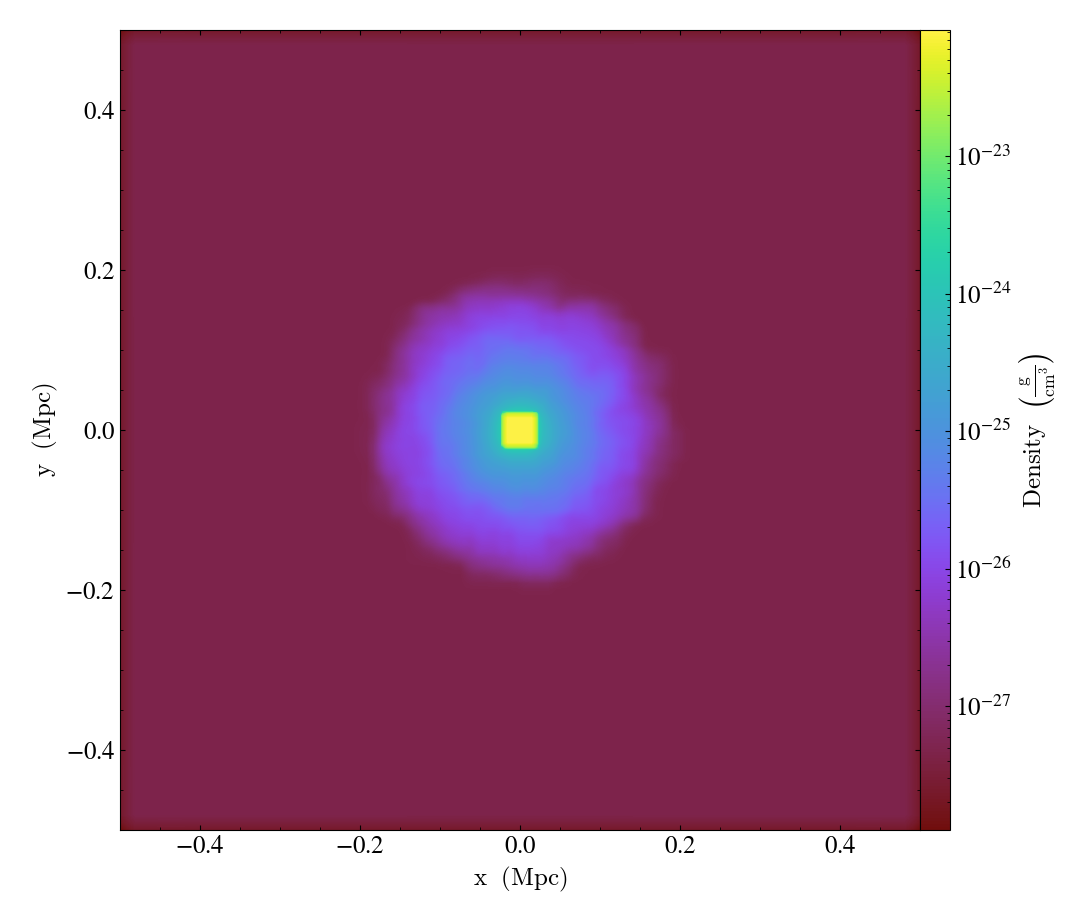
Applying filters on the final image¶
It is also possible to operate on the plotted image directly by using
one of the fixed resolution buffer filter as described in
Fixed Resolution Buffer Filters.
Note that it is necessary to call the plot object’s refresh method
to apply filters.
import yt
ds = yt.load('IsolatedGalaxy/galaxy0030/galaxy0030')
p = yt.SlicePlot(ds, 'z', 'density')
p.frb.apply_gauss_beam(sigma=30)
p.refresh()
p.save()
Extending annotations methods¶
New annotate_ methods can be added to plot objects at runtime (i.e., without
modifying yt’s source code) by subclassing the base PlotCallback class.
This is the recommended way to add custom and unique annotations to yt plots,
as it can be done through local plugins, individual scripts, or even external packages.
Here’s a minimal example:
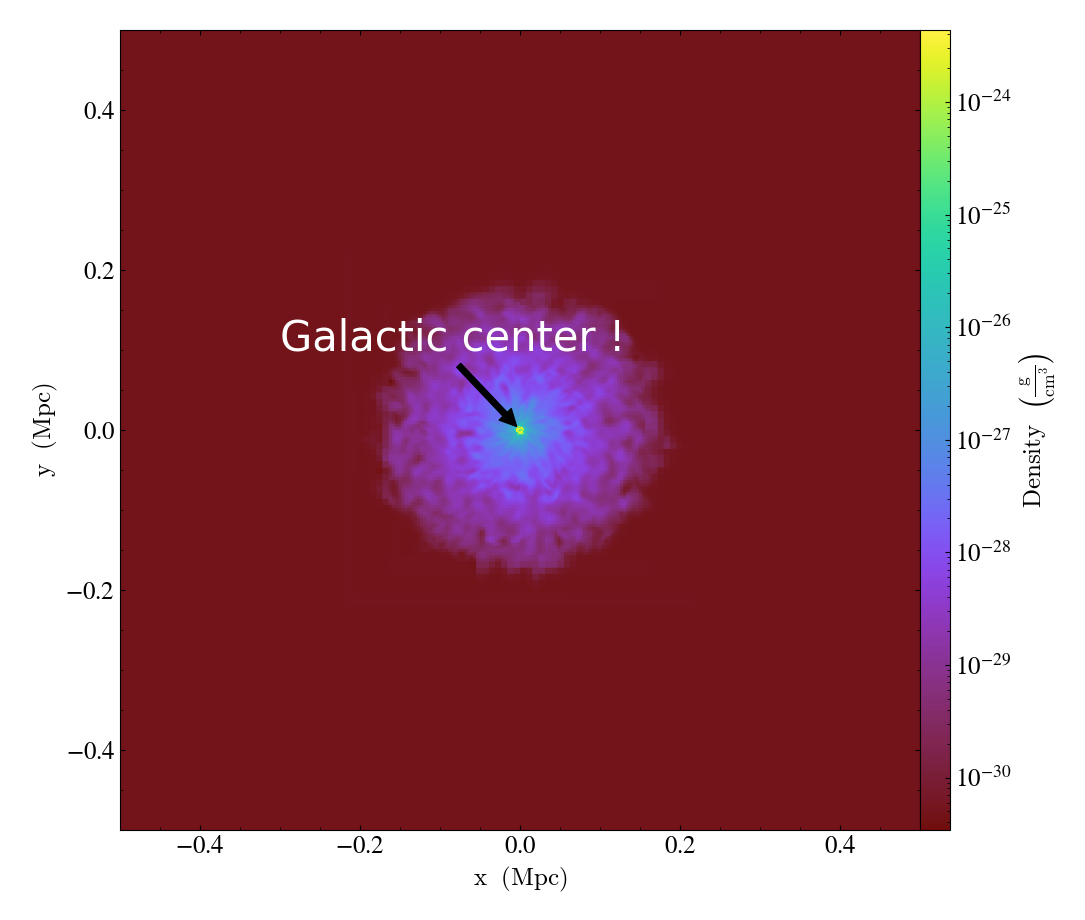
import yt
from yt.visualization.api import PlotCallback
class TextToPositionCallback(PlotCallback):
# bind a new `annotate_text_to_position` plot method
_type_name = "text_to_position"
def __init__(self, text, x, y):
# this method can have arbitrary arguments
# and should store them without alteration,
# but not run expensive computations
self.text = text
self.position = (x, y)
def __call__(self, plot):
# this method's signature is required
# this is where we perform potentially expensive operations
# the plot argument exposes matplotlib objects:
# - plot._axes is a matplotlib.axes.Axes object
# - plot._figure is a matplotlib.figure.Figure object
plot._axes.annotate(
self.text,
xy=self.position,
xycoords="data",
xytext=(0.2, 0.6),
textcoords="axes fraction",
color="white",
fontsize=30,
arrowprops=dict(facecolor="black", shrink=0.05),
)
ds = yt.load("IsolatedGalaxy/galaxy0030/galaxy0030")
p = yt.SlicePlot(ds, "z", "density")
p.annotate_text_to_position("Galactic center !", x=0, y=0)
p.save()