Particle Trajectories¶
One can create particle trajectories from a DatasetSeries object for a specified list of particles identified by their unique indices using the particle_trajectories method.
[1]:
%matplotlib inline
import glob
from os.path import join
import yt
from yt.config import ytcfg
path = ytcfg.get("yt", "test_data_dir")
import matplotlib.pyplot as plt
First, let’s start off with a FLASH dataset containing only two particles in a mutual circular orbit. We can get the list of filenames this way:
[2]:
my_fns = glob.glob(join(path, "Orbit", "orbit_hdf5_chk_00[0-9][0-9]"))
my_fns.sort()
And let’s define a list of fields that we want to include in the trajectories. The position fields will be included by default, so let’s just ask for the velocity fields:
[3]:
fields = ["particle_velocity_x", "particle_velocity_y", "particle_velocity_z"]
There are only two particles, but for consistency’s sake let’s grab their indices from the dataset itself:
[4]:
ds = yt.load(my_fns[0])
dd = ds.all_data()
indices = dd["all", "particle_index"].astype("int")
print(indices)
[1 2] dimensionless
which is what we expected them to be. Now we’re ready to create a DatasetSeries object and use it to create particle trajectories:
[5]:
ts = yt.DatasetSeries(my_fns)
# suppress_logging=True cuts down on a lot of noise
trajs = ts.particle_trajectories(indices, fields=fields, suppress_logging=True)
The ParticleTrajectories object trajs is essentially a dictionary-like container for the particle fields along the trajectory, and can be accessed as such:
[6]:
print(trajs["all", "particle_position_x"])
print(trajs["all", "particle_position_x"].shape)
[[0.25 0.25131274 0.25524942 0.26175659 0.27075597 0.28215068
0.2958247 0.31164457 0.32945507 0.3491025 0.37039668 0.39309658
0.41697977 0.44181091 0.46731412 0.49320738 0.51918241 0.54492553
0.57013802 0.5945386 0.61785738 0.63986883 0.66032111 0.67898584
0.69568959 0.71026758 0.72255999 0.73244151 0.73979734 0.7445359
0.74658248]
[0.75 0.74868726 0.74475058 0.7382434 0.72924402 0.71784928
0.70417524 0.68835534 0.67054478 0.65089727 0.62960299 0.60690298
0.58301967 0.5581884 0.53268507 0.50679169 0.48081655 0.45507332
0.42986072 0.40546004 0.38214117 0.36012962 0.33967725 0.32101242
0.30430854 0.28973042 0.27743785 0.26755617 0.26020016 0.25546143
0.25341467]] code_length
(2, 31)
Note that each field is a 2D NumPy array with the different particle indices along the first dimension and the times along the second dimension. As such, we can access them individually by indexing the field:
[7]:
plt.figure(figsize=(6, 6))
plt.plot(trajs["all", "particle_position_x"][0], trajs["all", "particle_position_y"][0])
plt.plot(trajs["all", "particle_position_x"][1], trajs["all", "particle_position_y"][1])
[7]:
[<matplotlib.lines.Line2D at 0x7f746ad1d090>]
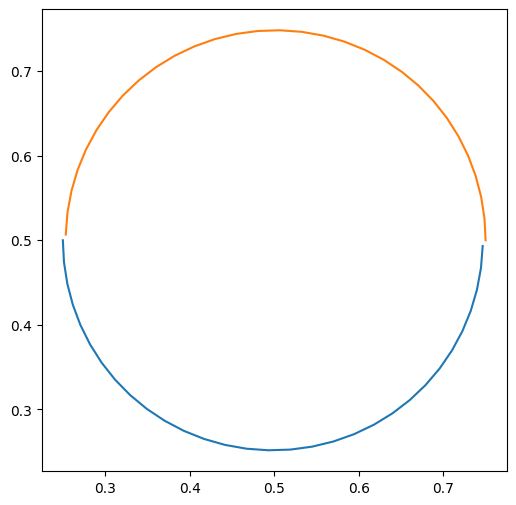
And we can plot the velocity fields as well:
[8]:
plt.figure(figsize=(6, 6))
plt.plot(trajs["all", "particle_velocity_x"][0], trajs["all", "particle_velocity_y"][0])
plt.plot(trajs["all", "particle_velocity_x"][1], trajs["all", "particle_velocity_y"][1])
[8]:
[<matplotlib.lines.Line2D at 0x7f746c5c7150>]
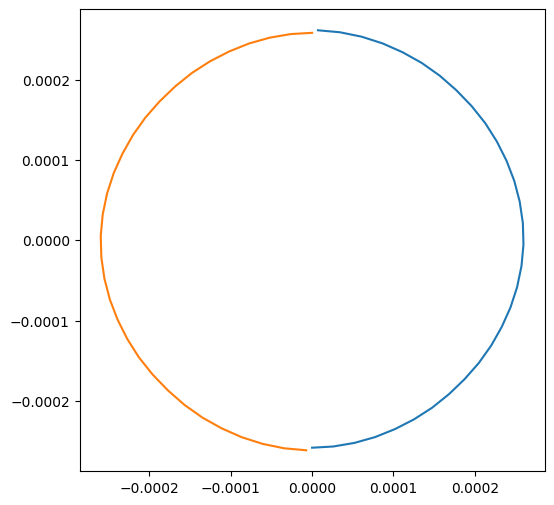
If we want to access the time along the trajectory, we use the key "particle_time":
[9]:
plt.figure(figsize=(6, 6))
plt.plot(trajs["particle_time"], trajs["particle_velocity_x"][1])
plt.plot(trajs["particle_time"], trajs["particle_velocity_y"][1])
[9]:
[<matplotlib.lines.Line2D at 0x7f746c556c50>]
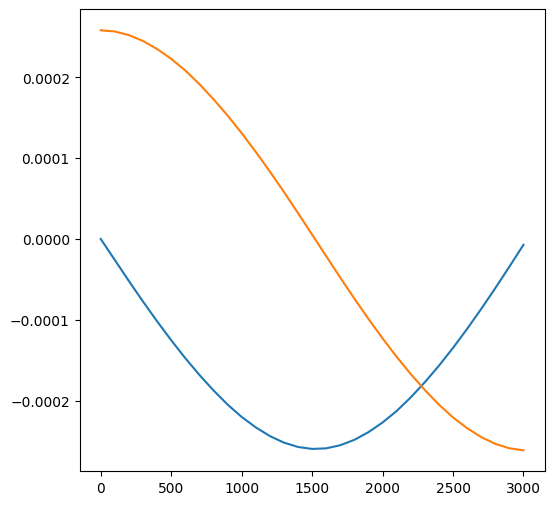
Alternatively, if we know the particle index we’d like to examine, we can get an individual trajectory corresponding to that index:
[10]:
particle1 = trajs.trajectory_from_index(1)
plt.figure(figsize=(6, 6))
plt.plot(particle1["all", "particle_time"], particle1["all", "particle_position_x"])
plt.plot(particle1["all", "particle_time"], particle1["all", "particle_position_y"])
---------------------------------------------------------------------------
TypeError Traceback (most recent call last)
Cell In[10], line 1
----> 1 particle1 = trajs.trajectory_from_index(1)
2 plt.figure(figsize=(6, 6))
3 plt.plot(particle1["all", "particle_time"], particle1["all", "particle_position_x"])
File /tmp/yt/yt/data_objects/particle_trajectories.py:340, in ParticleTrajectories.trajectory_from_index(self, index)
338 print("The particle index %d is not in the list!" % (index))
339 raise IndexError
--> 340 fields = sorted(self.field_data.keys())
341 traj = {}
342 traj[self.ptype, "particle_time"] = self.times
TypeError: '<' not supported between instances of 'str' and 'tuple'
Now let’s look at a more complicated (and fun!) example. We’ll use an Enzo cosmology dataset. First, we’ll find the maximum density in the domain, and obtain the indices of the particles within some radius of the center. First, let’s have a look at what we’re getting:
[11]:
ds = yt.load("enzo_tiny_cosmology/DD0046/DD0046")
slc = yt.SlicePlot(
ds,
"x",
[("gas", "density"), ("gas", "dark_matter_density")],
center="max",
width=(3.0, "Mpc"),
)
slc.show()
So far, so good–it looks like we’ve centered on a galaxy cluster. Let’s grab all of the dark matter particles within a sphere of 0.5 Mpc (identified by "particle_type == 1"):
[12]:
sp = ds.sphere("max", (0.5, "Mpc"))
indices = sp["all", "particle_index"][sp["all", "particle_type"] == 1]
Next we’ll get the list of datasets we want, and create trajectories for these particles:
[13]:
my_fns = glob.glob(join(path, "enzo_tiny_cosmology/DD*/*.hierarchy"))
my_fns.sort()
ts = yt.DatasetSeries(my_fns)
trajs = ts.particle_trajectories(indices, fields=fields, suppress_logging=True)
Matplotlib can make 3D plots, so let’s pick three particle trajectories at random and look at them in the volume:
[14]:
fig = plt.figure(figsize=(8.0, 8.0))
ax = fig.add_subplot(111, projection="3d")
ax.plot(
trajs["all", "particle_position_x"][100],
trajs["all", "particle_position_y"][100],
trajs["all", "particle_position_z"][100],
)
ax.plot(
trajs["all", "particle_position_x"][8],
trajs["all", "particle_position_y"][8],
trajs["all", "particle_position_z"][8],
)
ax.plot(
trajs["all", "particle_position_x"][25],
trajs["all", "particle_position_y"][25],
trajs["all", "particle_position_z"][25],
)
[14]:
[<mpl_toolkits.mplot3d.art3d.Line3D at 0x7f7466cfdd50>]
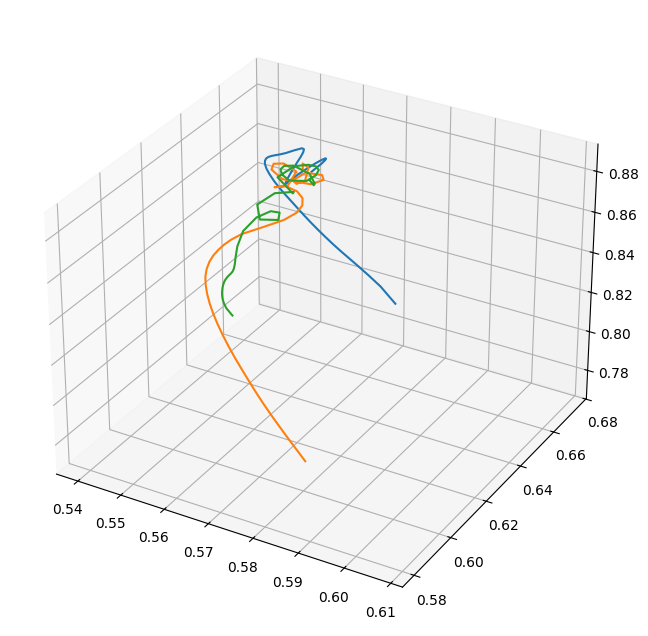
It looks like these three different particles fell into the cluster along different filaments. We can also look at their x-positions only as a function of time:
[15]:
plt.figure(figsize=(6, 6))
plt.plot(trajs["all", "particle_time"], trajs["all", "particle_position_x"][100])
plt.plot(trajs["all", "particle_time"], trajs["all", "particle_position_x"][8])
plt.plot(trajs["all", "particle_time"], trajs["all", "particle_position_x"][25])
---------------------------------------------------------------------------
YTFieldNotFound Traceback (most recent call last)
Cell In[15], line 2
1 plt.figure(figsize=(6, 6))
----> 2 plt.plot(trajs["all", "particle_time"], trajs["all", "particle_position_x"][100])
3 plt.plot(trajs["all", "particle_time"], trajs["all", "particle_position_x"][8])
4 plt.plot(trajs["all", "particle_time"], trajs["all", "particle_position_x"][25])
File /tmp/yt/yt/data_objects/particle_trajectories.py:162, in ParticleTrajectories.__getitem__(self, key)
160 return self.times
161 if key not in self.field_data:
--> 162 self._get_data([key])
163 return self.field_data[key]
File /tmp/yt/yt/data_objects/particle_trajectories.py:234, in ParticleTrajectories._get_data(self, fields)
232 new_particle_fields = []
233 for field in missing_fields:
--> 234 fds[field] = dd_first._determine_fields(field)[0]
235 if field not in self.particle_fields:
236 ftype = fds[field][0]
File /tmp/yt/yt/data_objects/data_containers.py:1471, in YTDataContainer._determine_fields(self, fields)
1468 explicit_fields.append(field)
1469 continue
-> 1471 finfo = self.ds._get_field_info(field)
1472 ftype, fname = finfo.name
1473 # really ugly check to ensure that this field really does exist somewhere,
1474 # in some naming convention, before returning it as a possible field type
File /tmp/yt/yt/data_objects/static_output.py:953, in Dataset._get_field_info(self, field)
948 def _get_field_info(
949 self,
950 field: FieldKey | ImplicitFieldKey | DerivedField,
951 /,
952 ) -> DerivedField:
--> 953 field_info, candidates = self._get_field_info_helper(field)
955 if field_info.name[1] in ("px", "py", "pz", "pdx", "pdy", "pdz"):
956 # escape early as a bandaid solution to
957 # https://github.com/yt-project/yt/issues/3381
958 return field_info
File /tmp/yt/yt/data_objects/static_output.py:1046, in Dataset._get_field_info_helper(self, field)
1043 elif (ftype, fname) in self.field_info:
1044 return self.field_info[ftype, fname], []
-> 1046 raise YTFieldNotFound(field, ds=self)
YTFieldNotFound: Could not find field ('all', 'particle_time') in DD0000.
Did you mean:
('all', 'particle_type')
<Figure size 600x600 with 0 Axes>
Suppose we wanted to know the gas density along the particle trajectory, but there wasn’t a particle field corresponding to that in our dataset. Never fear! If the field exists as a grid field, yt will interpolate this field to the particle positions and add the interpolated field to the trajectory. To add such a field (or any field, including additional particle fields) we can call the add_fields method:
[16]:
trajs.add_fields([("gas", "density")])
---------------------------------------------------------------------------
KeyError Traceback (most recent call last)
Cell In[16], line 1
----> 1 trajs.add_fields([("gas", "density")])
File /tmp/yt/yt/data_objects/particle_trajectories.py:212, in ParticleTrajectories.add_fields(self, fields)
197 def add_fields(self, fields):
198 """
199 Add a list of fields to an existing trajectory
200
(...)
210 >>> trajs.add_fields([("all", "particle_mass"), ("all", "particle_gpot")])
211 """
--> 212 self._get_data(fields)
File /tmp/yt/yt/data_objects/particle_trajectories.py:271, in ParticleTrajectories._get_data(self, fields)
269 for field in grid_fields:
270 pfield[field] = np.zeros(self.num_indices)
--> 271 x = self["particle_position_x"][:, step].d
272 y = self["particle_position_y"][:, step].d
273 z = self["particle_position_z"][:, step].d
File /tmp/yt/yt/data_objects/particle_trajectories.py:162, in ParticleTrajectories.__getitem__(self, key)
160 return self.times
161 if key not in self.field_data:
--> 162 self._get_data([key])
163 return self.field_data[key]
File /tmp/yt/yt/data_objects/particle_trajectories.py:302, in ParticleTrajectories._get_data(self, fields)
300 fd = fds[field]
301 for i, (_fn, (indices, pfield)) in enumerate(sorted(my_storage.items())):
--> 302 output_field[indices, i] = pfield[field]
303 self.field_data[field] = array_like_field(dd_first, output_field.copy(), fd)
305 if self.suppress_logging:
KeyError: 'particle_position_x'
We also could have included "density" in our original field list. Now, plot up the gas density for each particle as a function of time:
[17]:
plt.figure(figsize=(6, 6))
plt.plot(trajs["all", "particle_time"], trajs["gas", "density"][100])
plt.plot(trajs["all", "particle_time"], trajs["gas", "density"][8])
plt.plot(trajs["all", "particle_time"], trajs["gas", "density"][25])
plt.yscale("log")
---------------------------------------------------------------------------
YTFieldNotFound Traceback (most recent call last)
Cell In[17], line 2
1 plt.figure(figsize=(6, 6))
----> 2 plt.plot(trajs["all", "particle_time"], trajs["gas", "density"][100])
3 plt.plot(trajs["all", "particle_time"], trajs["gas", "density"][8])
4 plt.plot(trajs["all", "particle_time"], trajs["gas", "density"][25])
File /tmp/yt/yt/data_objects/particle_trajectories.py:162, in ParticleTrajectories.__getitem__(self, key)
160 return self.times
161 if key not in self.field_data:
--> 162 self._get_data([key])
163 return self.field_data[key]
File /tmp/yt/yt/data_objects/particle_trajectories.py:234, in ParticleTrajectories._get_data(self, fields)
232 new_particle_fields = []
233 for field in missing_fields:
--> 234 fds[field] = dd_first._determine_fields(field)[0]
235 if field not in self.particle_fields:
236 ftype = fds[field][0]
File /tmp/yt/yt/data_objects/data_containers.py:1471, in YTDataContainer._determine_fields(self, fields)
1468 explicit_fields.append(field)
1469 continue
-> 1471 finfo = self.ds._get_field_info(field)
1472 ftype, fname = finfo.name
1473 # really ugly check to ensure that this field really does exist somewhere,
1474 # in some naming convention, before returning it as a possible field type
File /tmp/yt/yt/data_objects/static_output.py:953, in Dataset._get_field_info(self, field)
948 def _get_field_info(
949 self,
950 field: FieldKey | ImplicitFieldKey | DerivedField,
951 /,
952 ) -> DerivedField:
--> 953 field_info, candidates = self._get_field_info_helper(field)
955 if field_info.name[1] in ("px", "py", "pz", "pdx", "pdy", "pdz"):
956 # escape early as a bandaid solution to
957 # https://github.com/yt-project/yt/issues/3381
958 return field_info
File /tmp/yt/yt/data_objects/static_output.py:1046, in Dataset._get_field_info_helper(self, field)
1043 elif (ftype, fname) in self.field_info:
1044 return self.field_info[ftype, fname], []
-> 1046 raise YTFieldNotFound(field, ds=self)
YTFieldNotFound: Could not find field ('all', 'particle_time') in DD0000.
Did you mean:
('all', 'particle_type')
<Figure size 600x600 with 0 Axes>
Finally, the particle trajectories can be written to disk. Two options are provided: ASCII text files with a column for each field and the time, and HDF5 files:
[18]:
trajs.write_out(
"halo_trajectories"
) # This will write a separate file for each trajectory
trajs.write_out_h5(
"halo_trajectories.h5"
) # This will write all trajectories to a single file
---------------------------------------------------------------------------
TypeError Traceback (most recent call last)
Cell In[18], line 1
----> 1 trajs.write_out(
2 "halo_trajectories"
3 ) # This will write a separate file for each trajectory
4 trajs.write_out_h5(
5 "halo_trajectories.h5"
6 ) # This will write all trajectories to a single file
File /tmp/yt/yt/utilities/parallel_tools/parallel_analysis_interface.py:364, in parallel_root_only.<locals>.root_only(*args, **kwargs)
361 @wraps(func)
362 def root_only(*args, **kwargs):
363 if not parallel_capable:
--> 364 return func(*args, **kwargs)
365 comm = _get_comm(args)
366 rv = None
File /tmp/yt/yt/data_objects/particle_trajectories.py:365, in ParticleTrajectories.write_out(self, filename_base)
348 @parallel_root_only
349 def write_out(self, filename_base):
350 """
351 Write out particle trajectories to tab-separated ASCII files (one
352 for each trajectory) with the field names in the file header. Each
(...)
363 >>> trajs.write_out("orbit_trajectory")
364 """
--> 365 fields = sorted(self.field_data.keys())
366 num_fields = len(fields)
367 first_str = "# particle_time\t" + "\t".join(fields) + "\n"
TypeError: '<' not supported between instances of 'str' and 'tuple'